A Comprehensive CRISPR-Cas9 Toolkit for Bacillus Subtilis: Development for Biomanufacturing Applications
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Mechanisms to the Human in Life Body
FROM DEFENSE BACTERIA MECHANISMS TO THE HUMAN IN LIFE BODY FOURTEENTH ANNUAL LSI SYMPOSIUM MAY 21, 2015 Images courtesy of Katherine D. Walton, research investigator, and Deborah Gumucio, professor, Department of Cell and Developmental Biology, U-M Medical School SCHEDULE FORUM HALL, PALMER COMMONS (OVERFLOW SEATING AVAILABLE IN GREAT LAKES NORTH) 8:30 A.M. 1:15 P.M. WELCOME | Alan Saltiel, Ph.D. ROLE OF THE MICROBIOTA IN INFECTION CONTROL Mary Sue Coleman Director of the Life Sciences Institute AND SEQUELAE | Yasmine Belkaid, Ph.D. Chief of Mucosal Immunology Section, Laboratory of 8:35 A.M. Parasitic Diseases, National Institutes of Health, National INTRODUCTION OF THE MARY SUE AND KENNETH Institute of Allergy and Infectious Diseases COLEMAN LIFE SCIENCES LECTURER | Mary Sue Coleman, Ph.D. 1:55 P.M. President Emerita IMMUNE REGULATION OF INTESTINAL HEALTH AND DISEASE | Gregory F. Sonnenberg, Ph.D. 8:50 A.M. Assistant Professor of Microbiology and Immunology in MARY SUE AND KENNETH COLEMAN LIFE SCIENCES Medicine, Weill Cornell Medical College LECTURE: HOMEOSTASIS, INFLAMMATION AND DISEASE | Ruslan Medzhitov, Ph.D. AFTERNOON BREAK David W. Wallace Professor of Immunobiology, Yale School of Medicine; Investigator, Howard Hughes 3:15 P.M. Medical Institute TISSUE CONTROL OF MACROPHAGE HOMEOSTASIS AND FUNCTION | Miriam Merad, M.D., Ph.D. MORNING BREAK Professor of Oncological Science and Medicine; Mount Sinai Chair in Cancer Immunology; Director of Human 10:30 A.M. Immune Monitoring Center, Tisch Cancer Institute, Icahn GENERATION OF A MEMORY OF INFECTION DURING School of Medicine at Mount Sinai CRISPR-CAS IMMUNITY | Luciano Marraffini, Ph.D. Assistant Professor, head of the Laboratory of 3:55 P.M. -

Patent-Lawyer-Article
The GLOBAL REACH, LOCAL KNOWLEDGE www.patentlawyermagazine.com Annual 2021 COPYRIGHT CTC LEGAL MEDIA The effect of design space on patent grant and recognition for designs Dr. Yongqiang Qi, Partner and Patent Attorney at Corner Stone, examines the latest judicial interpretation and what it means for design. Protect Patent validity against terms crises Page 56 Page 13 AI patenting Page 18 FFrontront ccover_TPL51_v2aover_TPL51_v2a Alternative.inddAlternative.indd 1 118/12/20208/12/2020 110:060:06 CRIPSR-Cas9 A Nobel Prize, a Global Pandemic, and a Patent Dispute walk into a bar… stop me if you’ve heard this one before Richard Gaugeler, Patent Attorney at Cedar White Bradley, explains how a Noble Prize, the Pandemic and a Patent Dispute are all inextricably linked to the CRISPR-Cas9 Technology. ust as the three individuals who walk into a The functioning of the CRISPR-Cas9 technology bar seemingly appear independent from is as follows: First, the faulty sequence in the Jone another, they nevertheless always turn DNA is identified by a scientist. In this case the out to be inextricably linked through some faulty sequence refers to a defective gene common thread. And our Nobel Prize, Pandemic, which codes for Sickle-Cell anaemia. Second, and Dispute are no different. The thread? Of course, CRISPR uses guideRNA to identify, and bind to I must be talking about CRISPR-Cas9 Technology. the sequence. The guideRNA binds to and The revolutionary gene editing tool that can be used unravels the faulty sequence in the DNA molecule. to make precise incisions in genetic material to Third, Cas9 cuts the faulty sequence to either edit or even delete unwanted genetic code. -

Convocation for Conferring Degrees Virtual Ceremony Thursday, June 11, 2020 Academic Procession New Castle Brass Quintet Welcomi
Convocation for Conferring Degrees Virtual Ceremony Thursday, June 11, 2020 Academic Procession New Castle Brass Quintet Welcoming Remarks Richard P. Lifton, M.D., Ph.D. President and Carson Family Professor Introduction Sidney Strickland, Ph.D. Dean of Graduate and Postgraduate Studies Vice President for Educational Affairs Conferring of the Degree of Doctor of Philosophy Dr. Lifton Presentation of the David Rockefeller Award for Extraordinary Service Dr. Lifton Alzatta Fogg Torsten N. Wiesel, M.D., F.R.S. Conferring of the Degree of Doctor of Science, Honoris Causa Dr. Lifton Marnie S. Pillsbury Lucy Shapiro, Ph.D. Academic Recession New Castle Brass Quintet 2 2020 Graduates Sarah Ackerman B.S., State University of New York, College at Geneseo The Role of Adipocytes in the Tumor Microenvironment in Obesity-driven Breast Cancer Progression Paul Cohen Sarah Kathleen Baker B.A., University of San Diego Blood-derived Plasminogen Modulates the Neuroimmune Response in Both Alzheimer’s Disease and Systemic Infection Models Sidney Strickland Mariel Bartley B.Sc., Monash University Characterizing the RNA Editing Specificity of ADAR Isoforms and Deaminase Domains in vitro Charles M. Rice Kate Bredbenner B.S., B.A., University of Rochester Visualizing Protease Activation, retroCHMP3 Activity, and Vpr Recruitment During HIV-1 Assembly Sanford M. Simon Ian Andrew Eckardt Butler B.A., The University of Chicago Hybridization in Ants Daniel Kronauer Daniel Alberto Cabrera* B.A., Columbia University Time-restricted Feeding Extends Longevity in Drosophila melanogaster Michael W. Young * Participant in the Tri-Institutional M.D.-Ph.D. Program 3 James Chen B.A., University of Pennsylvania Cryo-EM Studies of Bacterial RNA Polymerase Seth A. -

Bacteriophage-Based Synthetic Biology for the Study of Infectious Diseases
Bacteriophage-based synthetic biology for the study of infectious diseases The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Citorik, Robert J, Mark Mimee, and Timothy K Lu. “Bacteriophage- Based Synthetic Biology for the Study of Infectious Diseases.” Current Opinion in Microbiology 19 (June 2014): 59–69. As Published http://dx.doi.org/10.1016/j.mib.2014.05.022 Publisher Elsevier Version Final published version Citable link http://hdl.handle.net/1721.1/90322 Terms of Use Article is available under a Creative Commons license; see publisher's site for details. Detailed Terms http://creativecommons.org/ Available online at www.sciencedirect.com ScienceDirect Bacteriophage-based synthetic biology for the study of infectious diseases 1,2,5 1,2,5 1,2,3,4 Robert J Citorik , Mark Mimee and Timothy K Lu Since their discovery, bacteriophages have contributed Western World [5,6]. Though phages have remained an enormously to our understanding of molecular biology as important tool in the study of molecular biology, genetics, model systems. Furthermore, bacteriophages have provided and bacteria [7], concerns over the ever-dwindling arsenal many tools that have advanced the fields of genetic of antibiotics for the treatment of multidrug-resistant engineering and synthetic biology. Here, we discuss bacterial pathogens have also resulted in a renaissance bacteriophage-based technologies and their application to the in phage studies and in phage-based therapies as a means study of infectious diseases. New strategies for engineering to develop alternative therapeutics [8–11]. Correspond- genomes have the potential to accelerate the design of novel ingly, advances in synthetic biology have refined the phages as therapies, diagnostics, and tools. -

Determination of Fatty Acid Synthesis Intermediates in Escherichia Coli and Bacillus Subtilis
DETERMINATION OF FATTY ACID SYNTHESIS INTERMEDIATES IN ESCHERICHIA COLI AND BACILLUS SUBTILIS BY SWAMINATH SRINIVAS DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biochemistry in the Graduate College of the University of Illinois at Urbana-Champaign, 2018 Urbana, Illinois Doctoral Committee: Professor John E. Cronan Jr., Chair Professor James A. Imlay Professor Satish K. Nair Professor Wilfred A. van der Donk ABSTRACT Lipids play crucial roles in maintaining cellular structure and energy storage. Structural lipids in the form of phospholipids constitute almost 10% of the dry cell weight in bacteria with their synthesis requiring 32 moles of ATP per mole of lipid. This significant investment ensures that the flux through the fatty acid biosynthesis (FAS) and related metabolic pathways is very precisely coordinated. A key feature of the FAS pathway is the acyl carrier protein (ACP), which is a small acidic protein that tethers acyl intermediates via a high-energy thioester bond and shuttles them between enzymes. In Escherichia coli, ACP is one of the most abundant soluble proteins with about 60000 copies per cell. Despite being the subject of extensive biochemical and structural studies for several decades, a reliable snapshot of ACP-bound species in any organism under different conditions is unavailable. Previously used methods are severely limited in their capacity to differentiate fatty acid intermediates, suffer from poor reproducibility, require elaborate instrumentation and cannot be used in an ideal setting for determining intracellular fluxes. This dissertation describes a sensitive and facile method to identify and quantify the physiological level of acyl-ACP species in E. -

Research News … from Albany Medical Center
FALL 2017 Scientists studying cancer, Research News … cardiovascular disease, immunology and neurosciences help fulfi ll Albany Med’s mission as a major from Albany Medical Center biomedical research center. Quickening the Pace of Medical Discovery Albany Med’s research enterprise drives innovation in both patient care and education while also fueling the local economy and our reputation as a leader in developing new bioscientifi c knowledge and technology. The promise of biomedical and clinical research, INSIDE THIS ISSUE and the combination of the two, has never been greater. Gene Editing Pioneers With this newsletter we bring you up-to-date on some of the many exciting research 2 Selected to Receive activities at Albany Medical Center. Albany Prize Albany Med, Rensselaer Researchers 4 Collaborate to Advance Personalized Anti-Cancer Drugs Awards Assist Faculty 5 Researchers’ Pursuits Meet Our Newest 6 Faculty Members Please join us for a special Panel Discussion with the Pioneers of CRISPR-Cas9 Gene Editing, the 2017 recipients of the Albany Medical Center Prize in Medicine and Biomedical Research Tuesday, Sept. 26, 2017 4 - 5:30 p.m. Albany Medical Center ME-700 Light refreshments will be provided FALL 2017 Gene Editing Pioneers Selected to Receive Albany Prize For their roles in the creation of a remarkable gene Engineering, Massachusetts Institute of Technology, editing system that has been called the “discovery of the Cambridge, Mass. century,” fi ve researchers have been announced as the recipients of the Albany Medical Center Prize in Medicine The $500,000 award has been given annually since 2001 and Biomedical Research for 2017. -

From a Prokaryotic Immune System to a Gene Editing Tool Wenyan Jiang
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2016 CPISPR-CAS: From a Prokaryotic Immune System to a Gene Editing Tool Wenyan Jiang Follow this and additional works at: http://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons Recommended Citation Jiang, Wenyan, "CPISPR-CAS: From a Prokaryotic Immune System to a Gene Editing Tool" (2016). Student Theses and Dissertations. Paper 311. This Thesis is brought to you for free and open access by Digital Commons @ RU. It has been accepted for inclusion in Student Theses and Dissertations by an authorized administrator of Digital Commons @ RU. For more information, please contact [email protected]. CRISPR-CAS: FROM A PROKARYOTIC IMMUNE SYSTEM TO A GENOME EDITING TOOL A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Wenyan Jiang June 2016 © Copyright by Wenyan Jiang 2016 CRISPR-CAS: FROM A PROKARYOTIC IMMUNE SYSTEM TO A GENOME EDITING TOOL Wenyan Jiang, Ph.D. The Rockefeller University 2016 Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and their associated genes (cas) encode an adaptive, small-RNA-based immune system that protects prokaryotes from infectious phages and plasmids. CRISPR- Cas systems can be classified into three types based on their cas gene content. My thesis work focused on two parts. First, I investigated the mechanism and function of RNA cleavage in type III CRISPR-Cas immunity. Secondly, I developed a tool to manipulate prokaryotic genomes and gene expression by using an engineered type II CRISPR-Cas system. To date, all three types of CRISPR-Cas systems target DNA. -
CURATED CANCER CARE Physicians and Scientists in Oncoset Are Teaming up to Help Pioneer Precision Oncology
WINTER 2018 THE CRISPR REVOLUTION Northwestern Medicine scientists usher in a new era of genetic research • 16 INSIDE A REMARKABLE ONCOLOGY FULL SPECTRUM OF PRECISION YEAR • 10 CLOSE-UP • 20 GYNECOLOGIC CARE • 24 PATHOLOGIST • 28 FIRST GLANCE Northwestern Medicine Community Spotlight A Lighter Side of Medical School John Flaherty, MD, professor of Medicine JAMMING AT IN VIVO in the Division of Infectious Diseases, jams with second-year medical student Nick Volpe in a performance by “The Hypochondriacs” during the 39th annual production of In Vivo, Feinberg’s popular sketch comedy and variety show. Northwestern Medicine magazine Editorial Advisors: Eric G. Neilson, MD, Call or email us at 312-503-4210 or Connect with NM online: is published quarterly for alumni vice president for Medical Affairs and [email protected] fb.me/feinbergschoolofmedicine Lewis Landsberg Dean; Alan Krensky, ©2017 Northwestern University. and friends of Northwestern MD, vice dean for Development and Northwestern Medicine® is a federally twitter.com/nufeinbergmed University Feinberg School of Alumni Relations; Nicole Mladic, registered trademark of Northwestern flickr.com/feinbergschoolofmedicine Medicine, Northwestern Memorial executive director of Communications; Memorial HealthCare and is used by HealthCare and the McGaw Babette Nyka, director of Alumni Northwestern University. Don’t miss NM web extras! Relations Catch up on the latest Medical Center of Northwestern Material in Northwestern Medicine Northwestern Medicine news and University. Alumni Association: James P. Kelly, magazine may not be reproduced check out more photos and videos online ’73 MD, President; Rishi Reddy, ’00 MD, without prior consent and proper credit. at magazine.nm.org. Editor: Nora Dunne President-elect Address all correspondence to: Editorial Assistant: Yesenia Navarro Design: Taylor Design Northwestern University, Feinberg School Contributing Writers: Amber Bemis, of Medicine, Office of Communications Will Doss, Marla Paul, Cheryl SooHoo, 420 E. -
Patent Protection for CRISPR: an ELSI Review Jacob S
digitalcommons.nyls.edu Faculty Scholarship Articles & Chapters 2017 Patent Protection for CRISPR: An ELSI Review Jacob S. Sherkow New York Law School, [email protected] Follow this and additional works at: https://digitalcommons.nyls.edu/fac_articles_chapters Part of the Intellectual Property Law Commons, and the Science and Technology Law Commons Recommended Citation Journal of Law and the Biosciences, Vol. 4 (2017) This Article is brought to you for free and open access by the Faculty Scholarship at DigitalCommons@NYLS. It has been accepted for inclusion in Articles & Chapters by an authorized administrator of DigitalCommons@NYLS. Journal of Law and the Biosciences, 1-12 doi: 10.1093/jlb/lsx036 Advance Access Publication 0 2017 Essay Patent protection for CRISPR: An ELSI review Jacob S. Sherkow112 1. Innovation Center for Law and Technology, New York Law School, NY 10013, USA 2. Department of Health and Policy Management, Columbia University Mailman School of Public Health E-mail: [email protected] INTRODUCTION Much has been written about the power of CRISPR-the workhorse genetic-editing system first elucidated in 2012-and the public's interest in it, both as a piece of science and an ethical battleground.1 But there has also been extensive interest in the variety of intellectual property issues surrounding CRISPR, including a heated patent dispute between two of the technology's originators, Jennifer Doudna (UC Berkeley) and Emmanuelle Charpentier (Max-Planck), on one side, and Feng Zhang (Broad In stitute) on the other.2 While the intellectual property disputes concerning CRISPR are far from over-indeed, like Tolstoy's War and Peace, new characters central to the dis pute continue to materialize3 -five years of hindsight has given some perspective on their ethical, legal, and social implications. -
Sensing Danger COMMENTARY Luciano A
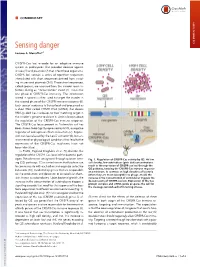
COMMENTARY Sensing danger COMMENTARY Luciano A. Marraffinia,1 CRISPR-Cas loci encode for an adaptive immune system in prokaryotes that provides defense against viruses (1) and plasmids (2) that infect these organisms. CRISPR loci contain a series of repetitive sequences intercalated with short sequences derived from invad- ing viruses and plasmids (3–5). These short sequences, called spacers, are acquired from the invader upon in- fection during an “immunization” event(1).Thisisthe first phase of CRISPR-Cas immunity. The information stored in spacers is then used to target the invader in the second phase of the CRISPR immune response (6). Each spacer sequence is transcribed and processed as a short RNA called CRISPR RNA (crRNA) that directs RNA-guided Cas nucleases to their matching target in the invader’s genome to cleave it. Little is known about the regulation of the CRISPR-Cas immune response. The CRISPR-Cas locus present in Escherichia coli has been shown to be tightly repressed by HNS, a negative regulator of widespread effects in bacteria (7). Repres- sion can be relieved by the LeuO activator (8), but en- vironmental or physiological conditions that lead to the expression of the CRISPR-Cas machinery have not been identified. In PNAS, Høyland-Kroghsbo et al. (9) describe the regulation of the CRISPR-Cas locus of the bacterial path- ogen Pseudomonas aeruginosa through quorum sens- Fig. 1. Regulation of CRISPR-Cas activity by QS. At low ing (QS) pathways. QS is a mechanism that bacteria use cell density, low autoinducer (pink dot) concentrations to communicate with each other and organize collective result in the repression of CRISPR-cas loci through the behaviors (10), mediated by genes that are responsible QS pathway, keeping the CRISPR-Cas immune response at a minimum. -

Center for Clinical and Translational Science Fall 2017
Fall 2017 Center for Clinical and Translational Science e- e-NewsletterNewsletter Center News Rockefeller University Biologist Michael W. Young Honored With Nobel Prize for Pioneering Studies on Circadian Rhythm The Rockefeller University website Dr. Young used genetics to identify his pioneering work on circadian rhythm,” gene mutations that disrupt the ability of says Richard P. Lifton, Rockefeller the fruit fly Drosophila melanogaster to University’s president. “The discoveries appropriately modulate its internal clock made by Mike and his colleagues have in response to a changing environment, provided fundamental insight into the and went on to define their biochemical molecular mechanisms by which the mechanisms. This clock found in fruit brain responds to environmental cues, a flies proves to be conserved throughout profound advance. The Nobel Prize is the the animal kingdom, and provides pinnacle of scientific recognition, and I insight into how the brain translates can think of no one more deserving than environmental cues into altered behavior. Mike to receive this award.” His work has direct implications for Dr. Barry Coller, Physician-in-Chief understanding human sleep disorders, of the Rockefeller University Hospital the mechanisms of jet lag, and the and Director of the Center for Clinical challenges of working on the night shift. and Translational Science (CCTS), noted Dr. Young’s lab is currently working “We are thrilled that the Nobel Prize 2017 Nobel Laureate and Rockefeller professor to assess how rhythmic gene and protein Committee is recognizing Mike for his Michael W. Young, Ph.D. activities are established in cells derived landmark studies with this singular from patients with sleep and depressive honor, and delighted that the CCTS, with Rockefeller University biologist disorders. -

CRISPR CAS9--LICENSING WHAT CAN't BE LICENSED Ulrich Storz | Les Nouvelles
CRISPR CAS9--LICENSING WHAT CAN'T BE LICENSED Ulrich Storz | les Nouvelles Document Details All Citations: 53 Les Nouvelles 123 Search Details Jurisdiction: National Delivery Details Date: July 9, 2018 at 12:40 AM Delivered By: kiip kiip Client ID: KIIPLIB02 Status Icons: © 2018 Thomson Reuters. No claim to original U.S. Government Works. CRISPR CAS9--LICENSING WHAT CAN'T BE LICENSED, 53 les Nouvelles 123 53 les Nouvelles 123 les Nouvelles June, 2018 Ulrich Storz a1 Copyright © 2018 by Licensing Executives Society International; Ulrich Storz CRISPR CAS9--LICENSING WHAT CAN'T BE LICENSED A new gene engineering technology has recently made it through the media, not only because of its technical advantages, but also because it is in the focus of an epic patent battle between two academic institutions. The technology bears the cryptic name “CRISPR Cas9,” and allows the manipulation of genes (so called “gene editing”) with so far unseen simplicity and efficacy. Dana Carroll of the University of Utah said for this reason that CRISPR Cas9 has brought about the “democratization of gene targeting.” 1 Probably, she made such statement also to work out a difference to a similar technology called Zinc Finger Nucleases, which Carrol has actively worked with, and which, besides being technically more challenging, were vigorously monopolized by a company called Sangamo Biosciences, by means of a proprietary library, and a restrictive patent licensing policy encompassing also non-commercial and academic use. 2 CRISPR Cas9 holds great promise not only for basic scientific research, but also for the generation of new plant varieties bearing advantageous features, for the generation of specific cell lines for cancer therapy, and even for the repair of fatally mutated genes in human embryos, to name a few potential applications.