Proceedings, No
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Influences of Squid Ink Added to Low Salt Fermented Squid on Its Changes in Lactic Acid Bacteria
Korean J. Food & Nutr. Vol. 26. No. 4, 678~684 (2013) THE KOREAN JOURNAL OF http://dx.doi.org/10.9799/ksfan.2013.26.4.678 한국식품영양학회지 FOOD AND NUTRITION Influences of Squid Ink Added to Low Salt Fermented Squid on Its Changes in Lactic Acid Bacteria †Sung-Cheon Oh Dept. of Food & Pharmacy, Daewon University College, Jecheon 390-702, Korea 저염 오징어 젓갈의 숙성 중 오징어 먹즙 첨가가 젖산균의 변화에 미치는 영향 †오 성 천 대원대학교 제약식품계열 국문요약 오징어 먹즙을 첨가하지 않은 저염 오징어 젓갈은 숙성 발효에 따라 숙성 초기에 젖산균 총수가 급속히 증가하고, Leuconostoc이 증가하여 숙성 적기에 최대량을 나타내고 이후 서서히 감소하였다. Lactobacillus는 숙성 적기 이후의 총 균수의 대부분을 차지할 만큼 균수가 많아지므로 숙성 적기 이후 품질에 관여하는 것으로 보인다. Streptococcus와 Pediococcus는 숙성 적기까지 완만하게 증가하다가 이후 급격히 감소하였다. Yeast는 숙성 중기 이후에 검출되고, 숙성 말기에 급격히 증가하므로 부패에 주로 관여하는 것으로 보인다. 오징어 먹즙을 첨가한 저염 오징어 젓갈의 숙성발효에 따른 젖산균의 변화를 보면 총 균수는 숙성 중반까지 증가하다가 중반 이후 약간 감소하였으나, 최대치에 도달하는 기간은 무 첨가군에 비하여 길었고, 젖산균 중 Leuconostoc, Streptococcus 및 Pediococcus는 숙성 후반까지 증가하였다가 후반에 감소하는 반면에, Lactobacillus는 숙성 후반까지 계속 증가하였다. Yeast는 숙성 초기에는 거의 증가하지 않다가 숙성 중반 이후 증가하였는데, 이러한 경향은 무 첨가군과 유사하였으나 젖산균의 수가 무 첨가군에 비하여 적었다. 오징어 먹즙을 첨가한 저염 오징어 젓갈에서 모든 균들의 수가 감소되어 오징어 먹즙이 균들의 성장 을 억제하는 것으로 확인되었다. Key words: lactic acid bacteria, low salt fermented squid, squid ink, yeast, ripening Introduction is a dominant strain in the ripening stage regardless of the salt concentration although higher salt concentration reduces the Fermented squid is a fermented seafood product which contains bacterial count in an extract (Mori et al. -

Cephalopod Gastronomy—A Promise for the Future
Cephalopod gastronomy - a promise for the future Mouritsen, Ole G.; Styrbæk, Klavs Published in: Frontiers in Communications DOI: 10.3389/fcomm.2018.00038 Publication date: 2018 Document version Publisher's PDF, also known as Version of record Document license: CC BY Citation for published version (APA): Mouritsen, O. G., & Styrbæk, K. (2018). Cephalopod gastronomy - a promise for the future. Frontiers in Communications, 3, [38]. https://doi.org/10.3389/fcomm.2018.00038 Download date: 27. Sep. 2021 REVIEW published: 29 August 2018 doi: 10.3389/fcomm.2018.00038 Cephalopod Gastronomy—A Promise for the Future Ole G. Mouritsen 1* and Klavs Styrbæk 2 1 Design and Consumer Behavior and Nordic Food Lab, Department of Food Science and Taste for Life, University of Copenhagen, Frederiksberg, Denmark, 2 STYRBÆKS, Odense, Denmark Cephalopods, specifically Coleoidea (squid, octopus, and cuttlefish), have for millennia been used as marine food by humans across the world and across different food cultures. It is particularly the mantle, the arms, the ink, and part of the intestines such as the liver that have been used. In addition to being consumed in the fresh and raw states, the various world cuisines have prepared cephalopods by a wide range of culinary techniques, such as boiling and steaming, frying, grilling, marinating, smoking, drying, and fermenting. Cephalopods are generally good nutritional sources of proteins, minerals, omega-3 fatty acids, as well as micronutrients, and their fat content is low. Whereas being part of the common fare in, e.g., Southeast Asia and Southern Europe, cephalopods are seldom used in regional cuisines in, e.g., North America and Northern Europe although the local waters there often have abundant sources of specific species that are edible. -

Pressed Sushi Sushi by Piece Rice Bowl Sushi Roll Sushi Combination
SUSHI BY PIECE RICE BOWL Bluefin Tuna 4.00 Salmon Don 16.00 Marinated Bluefin Tuna 4.00 Ikura Don 18.00 Kanpachi (Amberjack) 3.00 Sushi Hachi Chirashi 30.00 Hamachi (Yellow Tail) 3.00 Shimaaji (Stripedjack) 4.00 PRESSED SUSHI Madai (Seabream) 3.25 Saba Battera (6pcs) 20.00 Hirame (Flounder) 4.00 Aji (Jack Mackerel) 3.50 SUSHI COMBINATION Saba 3.50 Chef's Choice Sushi (10p+1r) 30.00 Sockeye Salmon 2.50 Ika (Squid) 2.50 SUSHI ROLL Tako (Octopus) 2.50 Amaebi (Raw Prawn) 3.50 Negi Toro (6pcs) 4.50 Tuna Belly & Green Onion Scallop 3.00 Mirugai (Geoduck) 4.00 California (6pcs) 4.50 Imitation Crab, Cucumber & Avocado Tamago (Egg) 2.00 Chopped Scallop 2.50 BC (5pcs) 6.00 Grilled Salmon Skin, Cucumber & Tobiko Spicy Scallop 2.50 Ikura (Salmon Roe) 3.00 Dynamite (5pcs) 6.00 Prawn Tempura, Avocado,Cucumber & Tobiko Tobiko 2.50 Bluefin Tuna Belly Daily House (4pcs) Daily Sashimi, Imitation Crab, 8.00 Grilled Salmon Skin, Cucumber, Tobiko & Prawn Tempura ***We accept VISA, Master & Debit card. ★Please pay at front. 8:30pm Last Order ☆GST not included. SASHIMI APPETIZER & SIDE ORDER Chef's Choice Sashimi 30.00 BBQ Bluefin Tuna 24.00 BBQ Salmon Neck 6.00 Kanpachi (Amberjack) 18.00 BBQ Madai Head 12.00~ Hamachi (Yellow Tail) 18.00 BBQ Shimaaji Head 15.00~ Shimaaji (Stripedjack) 24.00 BBQ Kanpachi Head 15.00~ Madai (Seabream) 18.00 Hirame (Flounder) 25.00 Aji (Jack Mackerel) 20.00 Others Saba (Mackerel) 20.00 Miso Soup 2.00 Sockeye Salmon 15.00 Rice 1.50 Ika (Squid) 12.00 Daikon Kinpira 3.00 Tako (Octopus) 12.00 Ankimo (Monk Fish Liver) 6.50 Amaebi (Raw Prawn) 21.00 Beef Sashimi 14.50 Scallop 18.00 Salmon Skin Salad 5.50 Mirugai (Geoduck) 24.00 Spicy Octopus Salad 8.50 Cooked Beef Tendon 5.50 Dessert Takowasa (Octopus & Wasabi) 4.50 Ice Cream 3.00 Japanese Hot Pot (Oden) 10.50 (Mango/Green Tea) Cooked Madai Head 12.50 8:30pm Last Order ***We accept VISA, Master & Debit card. -

Mycotoxins in Fermented Food1
[CANCER RESEARCH 28, 2296-2311, November 1968] Mycotoxins in Fermented Food1 R. Kinosita, T. Ishiko, S. Sugiyama, T. Seto, S. Igarasi, and I. E. Goetz City of Hope Medical Center, Duarte, California 91010 SUMMARY The biometrie statistics in Japan are quite reliable. The whole population is over 100 million, large enough to offer Selected for study fields were several distinct provincial reliable statistic figures. Moreover, practically all the people regions of Japan which are noted to be stable in population are of the same race, similar in habit and custom, literate, and composition, generally conservative in the mode of life, and live close to each other on the small well-developed land. distinguished by especially high figures in biometrie statistics Practically no one is left out of the official registration and representing the Japanese characteristics of extraordinarily census. According to a recent report issued from the Depart high incidence in death by cardiovascular diseases, stomach ment of Statistical Investigation, Japanese Ministry of Health cancer, and hepatoma. From these regions, samples of popular (Japanese Medical News, No. 2282, p. 108, 1968), the three fermented foodstuffs, consumed daily in quantity, were ob tained as materials of study. They included "miso" or fer highest causes of death in 1967 in Japan were apoplexy, mented soy bean paste, "katsuobushi" or fermented dry neoplasms, and heart diseases in that order, or more precisely, bonitos, and "starters" used for their fermentation processes. 25.6, 16.7, and 11.0 percent of the total death cases (about 670,000) respectively. The percentage of total death cases by They were either commercial products from local plants or cardiovascular diseases, including those by apoplexy and by home-made ones. -

Sashimi Tataki
sashimi & tataki Sashimi Moriawase 刺身盛り合わせ CupertinoDinnerserved all day assorted sashimi 28.00 Tako Sashimi たこ刺身 octopus 14.50 Sake Sashimi サーモン刺身 salmon 15.50 starters Madai Sashimi 真鯛刺身 red snapper 15.50 Edamame 枝豆 Hamachi Sashimi はまち刺身 lightly salted boiled soy beans 4.50 yellowtail 16.50 Hiyayakko やっこ Hirame Sashimi 平目刺身 cold tofu with ginger & scallions 4.50 halibut 16.50 Kimuchi キムチ Hirame Usuzukuri 平目うす造り korean spicy pickled cabbage 4.50 thinly sliced halibut with ponzu sauce 21.50 Shiokara 塩辛 Botan Ebi Sashimi ボタン海老刺身 salty fermented raw squid & innards 5.50 spot prawn 21.50 Chukafu Yakko 中華風やっこ Shiromaguro Tataki 白まぐろたたき cold tofu topped with seared albacore, garlic oil & ponzu 15.50 minced pork and vegetables 7.00 Yofu Yakko 洋風やっこ sliced tofu, tomatoes & shiso leaves carpaccio with garlic oil & balsamic ponzu vinaigrette 8.00 たこカルパッチョ Hiyashi Tomato 冷やしトマト Tako octopus, sliced cucumber, plum paste sliced chilled tomatoes 7.00 Tsukemono 漬物盛り合わせ vinaigrette dressing drizzled with garlic oil 16.50 assorted pickled vegetables 6.50 Salmon Aburi サーモンあぶりカルパッチョ Ebi Daikon えびの大根生春巻き balsamic-ponzu, grated shrimp, vegetables & mango rolled wasabi cucumber drizzled with truffle oil 17.50 with thinly sliced pickled daikon radish with Hirame 平目カルパッチョ a sweet vinaigrette sauce 9.00 halibut, shiso-pesto, & yuzu juice 18.50 ゆずセビッチェ Yuzu Ceviche Madai 真鯛カルパッチョ seafood of the day & yuzu quick-seared red snapper with salsa served with homemade potato chips 9.00 yuzu-pepper salsa 17.50 あん肝 Ankimo はまちカルパッチョ steamed monkfish liver in ponzu 10.50 -

Japan Update to 05.04.2021 Approval No Name Address Products Number FROZEN CHUM SALMON DRESSED (Oncorhynchus Keta)
Japan Update to 05.04.2021 Approval No Name Address Products Number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta). FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus). FROZEN JAPANESE SARDINE ROUND (Sardinops 81,Misaki-Cho,Rausu- Kaneshin Tsuyama melanostictus). FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma). 1 VN01870001 Cho, Menashi- Co.,Ltd FROZEN ALASKA POLLACK ROUND (Theragra chalcogramma). FROZEN PACIFIC COD Gun,Hokkaido,Japan DRESSED. (Gadus macrocephalus). FROZEN PACIFIC COD ROUND. (Gadus macrocephalus) Maekawa Shouten Hokkaido Nemuro City Fresh Fish (Excluding Fish By-Product); Fresh Bivalve Mollusk.; Frozen Fish (Excluding 2 VN01860002 Co., Ltd Nishihamacho 10-177 Fish By-Product); Frozen Processed Bivalve Mollusk; Frozen Chum Salmon(Round,Dressed,Semi-Dressed,Fillet,Head,Bone,Skin); Frozen 1-35-1 Alaska Pollack(Round,Dressed,Semi-Dressed,Fillet); Frozen Pacific Taiyo Sangyo Co.,Ltd. 3 VN01840003 Showachuo,Kushiro- Cod(Round,Dressed,Semi-Dressed,Fillet); Frozen Pacific Saury(Round,Dressed,Semi- Kushiro Factory City,Hokkaido,Japan Dressed); Frozen Chub Mackerel(Round,Fillet); Frozen Blue Mackerel(Round,Fillet); Frozen Salted Pollack Roe 3-9 Komaba- Taiyo Sangyo Co.,Ltd. 4 VN01860004 Cho,Nemuro- Frozen Fish ; Frozen Processed Fish; (Excluding By-Product) Nemuro Factory City,Hokkaido,Japan 3-2-20 Kitahama- Marutoku Abe Suisan 5 VN01920005 Cho,Monbetu- Frozen Chum Salmon Dressed; Frozen Salmon Dressed Co.,Ltd City,Hokkaido,Japan Frozen Chum Salmon(Round,Semi-Dressed,Fillet); Frozen Salmon Milt; Frozen Pink Salmon(Round,Semi-Dressed,Dressed,Fillet); -

APPETIZER / 前菜 Special Selected MINI HOMEMADE SOBA
BOWLSServed atop of Rice / 丼・重 Set Menu / Served with Miso Soup (Pork and Vegetables), Homemade Pickles and Steamed Egg Custard GYU SHIO KOJI DON 牛の塩麹焼き丼 APPETIZER / 前菜 Grilled and Sliced Chuck Roll Washu Beef marinated with SHIO KOJI (Salt-Marinated Rice Koji), Mashed Potato and Water Cress served with Wasabi A la carte 24 / Set 26 MAGURO YAMAKAKE 本マグロの山かけ うに添え OYAKO JU Blue Fin Tuna is served with Grated Japanese Grated Yam, Sea Urchin and Fried Bonito Flakes 親子重 16 Grilled Free Range Chicken and Onion is simmered in Dashi broth and later wrapped with layer of custard like Egg A la carte 16 / Set 18 HOMEMADE TOFU WITH WORLD SELECTED SALT 手作り豆腐 世界の塩で TORORO OYAKO DON Homemade Tofu served with Special Salt. Ask staff for detail as Salt is subject to change とろろ親子丼 6 Grilled Free Range Chicken and Onion is simmered in Dashi broth and later wrapped with layer of custard like Egg. Japanese Grated Yam is poured over the dish Special Selected A la carte 18 / Set 20 TORI KUROZU JU IKA SHIOKARA 自家製 いかの塩辛 鶏と野菜の黒酢あん重 Fried Chicken and Vegetables served with Ootoya famous “Sweet and Sour” Vinegar Sauce Sliced Raw Squid Cured in Salt and Squid Liver Marinade A la carte 16 / Set 18 7 EDAMAME YAKITORI JU 焼き鳥重 枝豆 世界の塩で Boiled Edamame sprinkled with Special Selected Salts Grilled Yakitori Assortments (Momo, Mune Wasabi, Tsukune, Tebamoto, Momo Asparagus) 6 A la carte 20 / Set 22 OHITASHI WITH SPECIAL BONITO FLAKES ROSU KATSU JU おひたし 本枯れ削り節添え 豚ロースかつ重 Boiled Green Vegetable and Shimeji served with Special Bonito Flakes. -
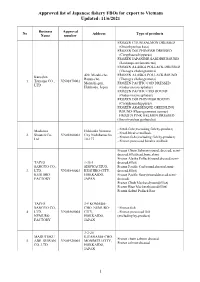
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN -

Summit Japanese Food Price List 30-4-2015
SUMMIT TRADING CO LLC , ABU DHABI , UAE - Tel + 971 2 6743100 WWW.SUMMIT.AE 冷凍日本食材リスト Apr.2015 JAPANESE FROZEN FOOD ITEMS Category Group Code Name Pcs/Ctn Packing 1 1 練り物 KB001 IKAMAKI 50 x 1 160 G Ground Fish Meat KB002 GOBOMAKI 50 x 1 160 G KB003 GOMOKUAGE 60 x 1 140 G KB004 KOBANAGE 60 x 1 120 G KB005 AKA KAMABOKO 100 x 1 160 G KB006 SHIRP KAMABOKO 100 x 1 160 G KB007 YAKI CHIKUWA 100 x 1 160 G KB008 CHIKUWA 60 x 1 160 G KB010 HANPEN 100 x 1 60 G KB014 AGEBALL 60 x 1 120 G KB015 EDAMAME CHIGIRI 60 x 1 140 G KB016 TAKOCHIGIRI 60 x 1 140 G KB017 IKARIKYU 60 x 1 120 G KB018 YASAI TSUMIRE 60 x 1 135 G KB019 KIBUN TSUMIRE 60 x 1 135 G KB020 UOGASHIAGE 60 x 1 140 G KB021 YASAI TEMPURA 60 x 1 120 G 92295 SURIMI 30 x 1 500 G 1 2 カニかまぼこ 91039 KANI KAMABOKO 20 x 2 500 G Kani Kamaboko 91074 KAORIBAKO 20 x 1 270 G KB009 KANI KAMABOKO 20 x 1 500 G KB025 SEA STIX 10 x 1 1,000 G KB026 SEA STIX 20 x 1 500 G 2 1 魚類 92077 GINDARA 5-7LBS/PCS 1 x 1 1,000 G Fish 92096 SANMA HIRAKI(SHINKU) 20 x 2 360 G 92102 SANMA ( 125G X 2 ) 30 x 1 125X2 G 92254 SHIRASU BOSHI 50 x 1 100 G 92299 HAMACHI (fillet) 7 x 1 1,000 G 92318 KAMAAGE SHIRASU 24 x 2 100 G 95370 HAZE HIRAKI 30 10 x 2 500 G 95372 KISU HIRAKI 30 10 x 2 600 G 95414 UNAGI KABAYAKI 1 x 2 5,000 G 95425 UNAGI KABAYAKI 1 x 1 5,000 G 95437 SABA SMOKE 20 x 2 100 G 95438 NISHIN SMOKE 20 x 1 80 G 95469 UNAGI KABAYAKI 50 x 2 80 G DT502 SHISHAMO 24 x 1 200 G DT506 UNAGI(Eel) KABAYAKI 45 x 1 203 G DT507 UNAGI(Eel) KABAYAKI 134 x 1 134 G DT513 AJINO HIRAKI 1 x 1 3,000 G DT539 SHIMESABA(Maackerel) 30 x 1 80 G DT540 -

Translation 4573
CANADIAN TRANSLATION OF FISHERIES AND AQUATIC SCIENCES No. 4573 Outline of the utilization of squid and octopus for processing by K. Miwa Original Title: Ika, tako no riyo kako no gaikyo From: Prog. Rep. Squid Fish. Survey World (5): 185-189, 1975 Translated by the Translation Bureau(ELC/PKW) Multilingual Services Division Department of the Secretary of State of Canada Department of Fisheries and Oceans Halifax Laboratory Halifax, N, S. 1979 16 pages typescript DEPARTMENT OF THE SECRETARY OF STATE SECRÉTARIAT D'ÉTAT TRANSLATION BUREAU ', BUREAU DES TRADUCTIONS MULTILINGUAL SERVICES -4Axw DIVISION DES SERVICES CANADA DIVISION MULTILINGUES C-i P'+5 #573 TRANSLATED FROM - TRADUCTION DE INTO - EN Japanese English AUTHOR - AUTEUR Katsutoshi MIWA TITLE IN ENGLISH - TITRE ANGLAIS Outline of the utilization of squid and octopus for processing TITLE IN FOREIGN LANGUAGE (TRANSLITERATE FOREIGN CHARACTERS) TITRE EN LANGUE ÉTRANGÉRE ( TRANSCRIRE EN CARACTÉRES ROMAINS) Ika, tako no riyo kako no gaikyo REFERENCE IN FOREIGN LANGUAGE (NAME OF BOOK OR PUBLICATION) IN FULL. TRANSLITERATE+FOREIGN CHARACTERS. RÉFÉRENCE EN LANGUE ÉTRANGÉRE (NOM DU LIVRE OU PUBLICATION),AU COMPLET, TRANSCRIRE EN CARACTÈRES ROMAINS. REFERENCE IN ENGLISH - REFERENCE EN ANGLAIS PUBLISHER- EDITEUR PAGE NUMBERS IN ORIGINAL DATE OF PUBLICATION NUMÉROS DES PAGES DANS DATE DE PUBLICATION L'ORIGINAL 185 - 189 YEAR ISSUE N0. VOLUME PLACE OF PUBLICATION ANNEE NUMERO NUMBER OF TYPED PAGES LIEU DE PUBLICATION NOMBRE DE PAGES DACTYLOGRAPHIÉES 16 2050679 REQUESTING DEPARTMENT DFO TRANSLATION BUREAU NO. MINISTÉRE-CLIENT NOTRE DOSSIER NO Sc. Info. & Pub. Br ELC /i KW BRANCH OR DIVISION TRANSLATOR (INITIALS) DIRECTION OU DIVISION-I VISION _ TRADUCTEUR ( INITIALES) Allan T. -

July Menu in Order to Provide Timely Food & Service, We Do Meiji Not Seat Parties Larger Than Ten
Izakayas are vibrant and casual Japanese pubs that serve drinks and small seasonal dishes that are meant to be shared July Menu In order to provide timely food & service, we do Meiji not seat parties larger than ten We serve all natural meat and locally grown produce when available To prepare our plates in a timely manner, we do not accommodate any menu substitutions 20% gratuity will be added to parties of six or more We charge 50 cents per item for to-go orders To-Go orders only taken in person “V” is for vegetarian, “GF” is for gluten free *Consuming raw or undercooked meats, poultry, Open daily 5pm to Late seafood, shellfish or eggs may increase your risk of foodborne illness Our restrooms are labeled “Water Closet” Kobachi Soup - Rice - Noodles (tiny dish) Wakame Salad 4 Land & Sea Donburi 6 V GF Seaweed salad, cucumber, dashi shoyu sauce Wakame, cucumber, red bell pepper, house-made rayu chili oil, steamed rice Japanese Pickles 3 V GF Curry Udon 12 House-made cucumber pickles Udon noodles, dashi broth, carrots, onion, thinly sliced beef, scallions Miso Egg* 3 V GF Zaru Soba 8 Marinated soft-boiled egg Japanese buckwheat noodle, roasted shiitake mentsuyu, green onion, grated Kimchi 4 GF daikon, fresh wasabi House-made napa cabbage Bay Scallops Tarako* 9 Squid ink pasta, mentaiko, bay scallops, parmesan butter, cream, shiso Edamame 4 V GF Soybeans, Korean roasted solar salt Shojin Chowder 6 V GF Rayu Edamame 5 V GF Turnips, snap peas, burdock, seasonal mushrooms Soybeans, house-made rayu chili oil, garlic Organic Miso Soup 4 GF Japanese -

The Whole Package a Couple's Lifetime of Love | The
The whole package: a couple's lifetime of love | The Japan Times Online 6/28/08 12:58 PM HOME The Japan Times Printer Friendly Articles MIXED MATCHES The whole package: a couple's lifetime of love By KAZUAKI NAGATA Staff writer In 1988, Yuki Takai, a native of Kyoto, and Jonathan Nordhausen from St. Paul, Minn., met in Atlanta while working for Yusen Air and Sea Service Co. Being in the delivery business, a lifetime of love and happiness might have been delivered to both Nordhausen, then an export manager, and Takai, who worked in sales, when they married in Georgia in 1994. International package: Yuki Takai and Jonathan Nordhausen pose with their daughter, Gretchen, at Takai's Their journey has now UPS office in Minato Ward, Tokyo, taken them to Tokyo, earlier this month. YOSHIAKI MIURA where they moved PHOTO from San Diego with their 13-year-old daughter, Gretchen, in January. Takai works as managing director for business development for United Parcel Service of America. Nordhausen, who has worked for several freight forwarders, takes care of their home in Japan. What did you find surprising or interesting about your partner's country? Yuki: The distance, like going shopping for 20 miles (32 km) is nothing (in the States). But in Japan, 30 km is a big deal. Back in the U.S., when I go to Los Angeles (from San Diego), it wasn't a big deal. It's an http://search.japantimes.co.jp/print/nn20080628f1.html Page 1 of 4 The whole package: a couple's lifetime of love | The Japan Times Online 6/28/08 12:58 PM Angeles (from San Diego), it wasn't a big deal.