Partitioning of the Maize Epigenome by the Number of Methyl Groups on Histone H3 Lysines 9 and 27
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Insights Into Hp1a-Chromatin Interactions
cells Review Insights into HP1a-Chromatin Interactions Silvia Meyer-Nava , Victor E. Nieto-Caballero, Mario Zurita and Viviana Valadez-Graham * Instituto de Biotecnología, Departamento de Genética del Desarrollo y Fisiología Molecular, Universidad Nacional Autónoma de México, Cuernavaca Morelos 62210, Mexico; [email protected] (S.M.-N.); [email protected] (V.E.N.-C.); [email protected] (M.Z.) * Correspondence: [email protected]; Tel.: +527773291631 Received: 26 June 2020; Accepted: 21 July 2020; Published: 9 August 2020 Abstract: Understanding the packaging of DNA into chromatin has become a crucial aspect in the study of gene regulatory mechanisms. Heterochromatin establishment and maintenance dynamics have emerged as some of the main features involved in genome stability, cellular development, and diseases. The most extensively studied heterochromatin protein is HP1a. This protein has two main domains, namely the chromoshadow and the chromodomain, separated by a hinge region. Over the years, several works have taken on the task of identifying HP1a partners using different strategies. In this review, we focus on describing these interactions and the possible complexes and subcomplexes associated with this critical protein. Characterization of these complexes will help us to clearly understand the implications of the interactions of HP1a in heterochromatin maintenance, heterochromatin dynamics, and heterochromatin’s direct relationship to gene regulation and chromatin organization. Keywords: heterochromatin; HP1a; genome stability 1. Introduction Chromatin is a complex of DNA and associated proteins in which the genetic material is packed in the interior of the nucleus of eukaryotic cells [1]. To organize this highly compact structure, two categories of proteins are needed: histones [2] and accessory proteins, such as chromatin regulators and histone-modifying proteins. -

Lecture No. VII Title of Topic: - Architecture of Chromosome Prepared By- Vinod Kumar, Assistant Professor, (PB & G) College of Agriculture, Powarkheda
Course: Fundamentals of Genetics Class: - Ist Year, IInd Semester Lecture No. VII Title of topic: - Architecture of Chromosome Prepared by- Vinod Kumar, Assistant Professor, (PB & G) College of Agriculture, Powarkheda Introduction:- E. Strasburger in 1875 first discovered thread-like structures which appeared during cell division. These thread like structures were called chromosomes due to their affinity for basic dyes. The term chromosome is derived from two Greek words; chrom = colour, soma=body. This term was first used by Waldeyer in 1888. Morphology: The outer covering or sheath of a chromosome is known as pellicle, which encloses the matrix. Within the matrix lies the chromatin. Flemming introduced the term chromatin in 1879. The term chromatin refers to the Feulgen positive materials observed in interphase nucleus and later during nuclear division. Chromatin readily stains with basic dyes especially Basic Fuchsin, which is specific for DNA which in turn is a major constituent of chromosomes. The chromosome morphology changes during cell division and mitotic metaphase is the most suitable stage for studies on chromosome morphology. In mitotic metaphase chromosomes, the following structural features can be seen under the light microscope. 1. Chromatid: Each metaphase chromosome appears to be longitudinally divided into two identical parts each of which is called chromatid. Both the chromatids of a chromosome appear to be joined together at a point known as centromere. The two chromatids of chromosome separate from each other during mitotic anaphase (and during anaphase II of meiosis) and move towards opposite poles. Since the two chromatids making up a chromosome are produced through replication of a single chromatid during synthesis (S) phase of interphase, they are referred to as sister chromatids. -

Mapping Epigenetic Modifications on Chicken Lampbrush Chromosomes Tatiana Kulikova , Anna Surkova , Anna Zlotina and Alla Krasikova*
Kulikova et al. Molecular Cytogenetics (2020) 13:32 https://doi.org/10.1186/s13039-020-00496-0 RESEARCH Open Access Mapping epigenetic modifications on chicken lampbrush chromosomes Tatiana Kulikova , Anna Surkova , Anna Zlotina and Alla Krasikova* Abstract Background: The epigenetic regulation of genome is crucial for implementation of the genetic program of ontogenesis through establishing and maintaining differential gene expression. Thus mapping of various epigenetic modifications to the genome is relevant for studying the regulation of gene expression. Giant transcriptionally active lampbrush chromosomes are an established tool for high resolution physical mapping of the genome and its epigenetic modifications. This study is aimed at characterizing the epigenetic status of compact chromatin domains (chromomeres) of chicken lampbrush macrochromosomes. Results: Distribution of three epigenetic modifications – 5-methylcytosine, histone H3 trimethylated at lysine 9 and hyperacetylated histone H4 – along the axes of chicken lampbrush chromosomes 1–4, Z and W was analyzed in details. Enrichment of chromatin domains with the investigated epigenetic modifications was indicated on the cytological chromomere-loop maps for corresponding chicken lampbrush chromosomes. Heterogeneity in the distribution of 5-methylcytosine and histone H3 trimethylated at lysine 9 along the chromosome axes was revealed. Conclusions: On examples of certain chromomeres of chicken lampbrush chromosomes 1, 3, 4 and W we demonstrated that a combination of immunofluorescent staining and fluorescence in situ hybridization allows to relate the epigenetic status and a DNA sequence context of individual chromomeres. Keywords: Chromatin domain, Chromomere, Cytological chromomere-loop map, Chicken, Epigenetic modifications, Gene mapping, Histone modifications, Karyotype, Lampbrush chromosomes, Methylated cytosine, Tandem repeats Background active chromatin is organized in paired lateral loops out- Lampbrush chromosomes are highly extended transcrip- going from the chromomeres. -
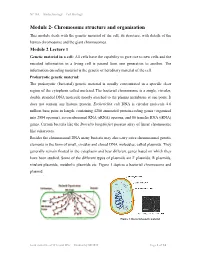
Chromosome Structure and Organisation
NPTEL – Biotechnology – Cell Biology Module 2- Chromosome structure and organisation This module deals with the genetic material of the cell, its structure, with details of the human chromosome and the giant chromosomes. Module 2 Lecture 1 Genetic material in a cell: All cells have the capability to give rise to new cells and the encoded information in a living cell is passed from one generation to another. The information encoding material is the genetic or hereditary material of the cell. Prokaryotic genetic material: The prokaryotic (bacterial) genetic material is usually concentrated in a specific clear region of the cytoplasm called nucleiod. The bacterial chromosome is a single, circular, double stranded DNA molecule mostly attached to the plasma membrane at one point. It does not contain any histone protein. Escherichia coli DNA is circular molecule 4.6 million base pairs in length, containing 4288 annotated protein-coding genes (organized into 2584 operons), seven ribosomal RNA (rRNA) operons, and 86 transfer RNA (tRNA) genes. Certain bacteria like the Borrelia burgdorferi possess array of linear chromosome like eukaryotes. Besides the chromosomal DNA many bacteria may also carry extra chromosomal genetic elements in the form of small, circular and closed DNA molecules, called plasmids. They generally remain floated in the cytoplasm and bear different genes based on which they have been studied. Some of the different types of plasmids are F plasmids, R plasmids, virulent plasmids, metabolic plasmids etc. Figure 1 depicts a bacterial chromosome and plasmid. Figure 1: Bacterial genetic material Joint initiative of IITs and IISc – Funded by MHRD Page 1 of 24 NPTEL – Biotechnology – Cell Biology Virus genetic material: The chromosomal material of viruses is DNA or RNA which adopts different structures. -

Lampbrush Chromosomes
COMMENTARY Lampbrush chromosomes H. C. MACGREGOR Department of'/.oology, Univeisity of Leicester, Leicester LEI 7RH, UK 1986 was a celebration year for lampbrush chromo- problems of the fate and significance of the massive somes (LBCs), marked by the publication of Callan's amounts of non-coding transcripts, and the manner in comprehensive and authoritative book on these struc- which certain initiation sites are 'selected' for high-level tures. My commentary begins where Callan's book transcription on lateral loops, whereas others remain ends, standing on tiptoe beside a large and rather relatively inactive on or within the chromomeres. Of neat assembly of well-established facts, principles and particular interest are the recent studies of Epstein et hypotheses, and trying hard to catch a glimpse of what al. (1986) and Epstein & Gall (1987). These investi- lies ahead. gators have focussed their attention on the nature and Several major principles can now be accepted with transcription of the 330 base-pair (bp) satellite DNA of confidence. LBCs are a feature of the growing oocytes Notophthalmus viridescens (Nv2). Nv2 occurs in tan- of most animals, except mammals and certain insects. demly repeated clusters throughout the genome and its Callan (1986) has reviewed this principle in depth, and sequence is highly conserved among the salamandrid has discussed the significance of exceptional cases. The species so far examined (Epstein et al. 1986). It is main defining feature of LBCs, their lateral loops, are transcribed on lampbrush loops by read-through from regions of intense RNA synthesis, and together these adjacent structural gene promoters, and homologous loops produce a large variety of transcription products, strand-specific cytoplasmic transcripts are found in the though not all coding sequences in the genome are cytoplasm of a variety of tissues. -

Visualization of Early Chromosome Condensation: a Hierarchical Folding, Axial Glue Model of Chromosome Structure
JCBArticle Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure Natashe Kireeva,1 Margot Lakonishok,1 Igor Kireev,1 Tatsuya Hirano,2 and Andrew S. Belmont1 1Department of Cell and Structural Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801 2Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724 urrent models of mitotic chromosome structure are appear until late prophase, after formation of uniformly based largely on the examination of maximally con- condensed middle prophase chromosomes. Instead, SMC2 C densed metaphase chromosomes. Here, we test these associates throughout early and middle prophase chroma- models by correlating the distribution of two scaffold com- tids, frequently forming foci over the chromosome exterior. ponents with the appearance of prophase chromosome Early prophase condensation occurs through folding of folding intermediates. We confirm an axial distribution of large-scale chromatin fibers into condensed masses. These topoisomerase II␣ and the condensin subunit, structural resolve into linear, 200–300-nm-diameter middle prophase maintenance of chromosomes 2 (SMC2), in unextracted chromatids that double in diameter by late prophase. We metaphase chromosomes, with SMC2 localizing to a 150– propose a unified model of chromosome structure in which 200-nm-diameter central core. In contrast to predictions of hierarchical levels of chromatin folding are stabilized late radial loop/scaffold models, this axial distribution does not in mitosis by an axial “glue.” Introduction Understanding the structural and molecular basis of mitotic predictions in terms of the functional mechanisms under- chromosome condensation remains a basic challenge in cell lying chromosome condensation and the types of folding biology. -

A Method for Determination of the in Situ Distribution of Chromosomal Proteins
Proc. Nat. Acad. Sci. USA Vol. 73, No. 2, pp. 423-427, February 1976 Biochemistry A method for determination of the in situ distribution of chromosomal proteins (nonhistone chromosomal proteins/chromosome structure/Drosophila melanogaster/polytene chromosomes/immunofluorescence) LEE M. SILVER* AND SARAH C. R. ELGINt Committee on Higher Degrees in Biophysics and t Department of Biochemistry and Molecular Biology, The Biological Laboratories, Harvard University, Cambridge, Massac usetts 02138 Communicated by Matthew S. Meselson, December 1, 1975 ABSTRACT A technique has been developed for "stain- tracted from chromatin with 1.6 M NaCl-0.2 M HC1. The ing" cytological preparations by indirect immunofluorescent DNA and NHC proteins were solubilized in 0.05 M Tris, pH methods that permits determination of the in situ distribu- tion of chromosomal proteins. The method is particularly ori- 8-1% sodium dodecyl sulfate (NaDodSO4), and the DNA ented to the use of polytene chromosome squashes from Dro- was removed by centrifugation (3). The supernatant con- sophila salivary glands. Control experiments indicate that tains the NHC proteins; this preparation will be referred to the fixation methods used allow little or no extraction or re- as NHCP-1. (ii) Chromatin was dissociated in 5 M urea-2 M arrangement of the chromosomal proteins. The results ob- NaCl-0.001 M sodium phosphate buffer, pH 7.0-1 mM so- tained demonstrate the specific in vivo chromosomal loca- tions of nonhistone proteins purified from isolated chroma- dium bisulfite. The histones, NHC proteins, and DNA were tin. The technique is apparently capable of resolution at the then eluted sequentially from a hydroxylapatite column level of the chromomere or band, the unit of genetic organi- with 0.001, 0.1, and 0.5 M phosphate (4). -

Chromomeres Revisited
Chromosome Res DOI 10.1007/s10577-012-9310-3 Chromomeres revisited Herbert C. Macgregor # Springer Science+Business Media B.V. 2012 Abstract The history of studies on the chromomeres Keywords lampbrush chromosome . chromomeres . of lampbrush chromosomes is outlined and evidence chromatin . transcription for the nature and function of these structures is col- lected and summarised. Chromomeres and their asso- Abbreviations ciated loops on lampbrush chromosomes are not bp Base pairs genetic units although in some special cases, they CENP-A Centromeric protein A consist of specific families of repeated DNA sequen- DAPI 4′,6-Diamidino-2-phenylindole ces. The emergence of a chromomeric organisation LBC Lampbrush chromosome coincides with the onset and intensification of tran- H4 Histone H4 scription on lampbrush loops. Modern molecular stud- H3K4me3 A modified histone H3 that marks ies have provided evidence that the chromatin of transcriptional activation lampbrush chromomeres differs in several important HMGN High molility group nucleosome binding respects from that of condensed metaphase chromo- proteins somes. It is in a highly dynamic state that facilitates HP A protein involved in heterochromatin localised transcription whilst keeping the chromosome formation and gene silencing safe from structural changes that might impede its SMC Structural maintenance of chromosome orderly progression up to and through meiotic meta- protein phase 1. Lampbrush chromosomes (LBCs) are a phys- RAD21 A protein involved in chromosome ically induced phenomenon, facilitated by the condensation in mitosis selective absence of molecular factors that would XCAP-D2 A condensin complex with a role in interfere with their main transcriptional role. LBC chromosome condensation and segregation morphology is highly dynamic and driven by tran- scriptive activity. -

Classification of Loops of Lampbr Ush Chromosomes According to the Arrangement of Transcriptional Complexes
J. Cell Sei. 22, 503-519 (1976) Printed in Great Britain CLASSIFICATION OF LOOPS OF LAMPBR USH CHROMOSOMES ACCORDING TO THE ARRANGEMENT OF TRANSCRIPTIONAL COMPLEXES U. SCHEER, W. W. FRANKE, M. F. TRENDELENBURG AND H. SPRING Division of Membrane Biology and Biochemistry, Institute of Experimental Pathology, German Cancer R esearch Center, D-69 Heidelberg, Federal Republic of Germany SUMMARY The arrangement of transcriptional units in the loops of lampbrush chromosomes from oocyte nuclei of urodele amphibia and from primary nuclei of the green alga Acetabularia have been studied in the electron microscope using spread preparations. Loops with different patterns of arrangement of matrix units (i.e. to a first approximation, transcriptional units) can be distinguished: (i) loops consisting of one active transcriptional unit; (ii) loops containing one active transcriptional unit plus additional fibril-free, i. e. apparently untranscribed, inter cepts that may include 'spacer' regions; (iii) loops containing two or more transcriptional units arranged in identical or changing polarities, with or without interspersed apparent spacer regions. Morphological details of the transcriptional complexes are described. The observations are not compatible with the concept that one loop reflects one and only one transcriptional unit but, rather, lead to a classification of loop types according to the arrangement of their transcriptional units. We propose that the lampbrush chromosome loop can represent a unit for the coordinate transcription of either one gene or a set of several (different) genes. TRODUCTION Lampbrush chromosomes offer the unique possibility of studying structural aspects of transcription within individual, cytologicaUy defined subunits of chromo somes, the 'chromomeres' (for definition see the recent review by Lima-de-Faria, 1975). -

Avian Lampbrush Chromosomes: a Powerful Tool for Exploration of Genome Expression
Review Article Cytogenet Genome Res 2009;124:251–267 Accepted after revision: October 14, 2008 DOI: 10.1159/000218130 by A. Houben Avian Lampbrush Chromosomes: a Powerful Tool for Exploration of Genome Expression E. Gaginskaya T. Kulikova A. Krasikova Saint-Petersburg State University, Saint-Petersburg, Russia Key Words some specific features: very long transcription units, deregu- Chicken genome ؒ C h r o m o s o m e o r g a n i z a t i o n ؒ L a m p b r u s h lated termination, and transcription of non-coding satellite chromosomes ؒ Non-coding RNA ؒ Oocyte nucleus ؒ repeats. Here, based on the modern view on a role of RNA ,Oogenesis ؒ T r a n s c r i p t i o n interference machinery in regulation of genome expression we suggest a mechanism of initiation of satellite DNA tran- scription and offer a novel interpretation of the ‘classical’ hy- Abstract pothesis that sought to explain the significance of wide- Lampbrush chromosomes (LBCs) are highly extended biva- spread transcription during oocyte growth. lents that function in the growing oocytes of many animals. Copyright © 2009 S. Karger AG, Basel Due to their distinctive chromomere-loop organization and intense transcriptional activity of lateral loops the LBCs, mainly amphibian ones, have served as a powerful system The investigation of genome structure and function for exploring the general principles of chromosome organi- has been greatly helped by studies of model systems such zation and function. The exploitation of avian LBCs has con- as polytene and lampbrush chromosomes that facilitate siderably broadened the opportunities for comparative exploration of gene activity in vivo. -

Distribution of Chromomeres As a Basis of Chromosomal Coiling
J. Cell Sd. 80, 193-205 (1986) 193 Printed in Great Britain © The Company of Biologist! Limited 1986 DISTRIBUTION OF CHROMOMERES AS A BASIS OF CHROMOSOMAL COILING VEIKKO SORSA Department of Genetics, University of Helsinki, Finland SUMMARY Periodicity in the distribution of prominent bands was analysed from the light and electron microscopic maps of salivary gland chromosomes of Drosophila melanogaster. The data obtained indicate that a similar distribution of prominent chromomeres in an individual interphase chromatid results in a unilateral accumulation of chromatin at the chromonema stage, if the helical axis of chromonema consists of ~S-9 interchromomere + chromomere units per turn. Orientation of the largest chromomeres mainly on one lateral half and the smallest chromomeres mairrly on the opposite lateral half of the chromonema apparently bends it to form the chromosomal 'macro* coil. Thus the increase in DNA content in the chromomeric loops located at specific intervals along the chromatids may have an important role in the evolution of coiling hierarchy in the eukaryotic chromosomes. INTRODUCTION At about the same time in the last century as the helical coiling of chromosomes was first depicted by Baranetzky (1880) the polytene chromosomes were also described as striated cords in the cell nuclei of certain insects (Balbiani, 1881). Since then an abundance of reports has been published concerning both the coiling of chromosomes (see e.g. Kaufmann, 1936, 1948; Ris, 1961; Ris & Korenberg, 1979) and the banding pattern in the polytenized interphase chromosomes (see Beermann, 1962). Although chromosomal coiling has been clearly demonstrable by means of light microscopy (LM) (e.g. see Ohnuki, 1968), electron microscopy (EM) of whole mounts has often failed to uncover it, and favoured the folded fibre or radial loop models of mitotic chromosomes instead of coiling hierarchy (DuPraw, 1970; Labhart, Koller & Wunderli, 1982; Utsumi, 1982). -

The Mitotic Chromosome: Structure and Mechanics John F
| 449 18 The Mitotic Chromosome: Structure and Mechanics John F. Marko 18.1 Introduction Cell division requires replication of DNA followed by physical segregation of chromosomes and replicated chromatids. DNA replication is likely to leave duplicate sister chromatids wrapped around one another (catenated) at least once every few kilobases (Chapter 15). Then, as the cell enters mitosis, chromosomes are folded (“condensed”) into neatly compacted and segregated mitotic chromo- somes each composed of two separated sister chromatids (Figure 18.1). The mechanism of mitotic folding likely facilitates segregation of chromosomes and adjacent chromatids, but exactly how this occurs – both the folding and its cou- pling to topological and geometrical segregation – is largely a mystery [1]. Our incomplete understanding of chromosome organization follows in part from basic physical properties of chromatin. The dynamic nature of chromosomes resulting from active processes including gene positioning and cell cycle reorga- nization, plus continual thermal (Brownian) motions indicates that chromatin structure must be described statistically, rather than in terms of precise folds and structures. Furthermore, chromatin and chromosomes are soft materials, with rigidities far less than that of the molecules from which they are composed, leading to the pitfall that large-scale structure of chromosomes can be altered by preparations which leave protein and DNA molecular structures intact. Finally, the molecular mechanisms by which large-scale chromosome organization is con- trolled are simply not understood. This chapter focuses on the large-scale architecture, mechanics, and molecular connectivity of mitotic chromosomes. Section 18.2 reviews protein components thought to define the structure of mitotic chromosomes, starting with histones and other key DNA-binding proteins, and then examining the “structural main- tenance of chromosomes” (SMC) protein complexes known to be essential to formation and maintenance of mitotic chromosome structure [2–5].