Is There Any Intron Sliding in Mammals? Irina V
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

The Landscape of Genomic Imprinting Across Diverse Adult Human Tissues
Downloaded from genome.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Research The landscape of genomic imprinting across diverse adult human tissues Yael Baran,1 Meena Subramaniam,2 Anne Biton,2 Taru Tukiainen,3,4 Emily K. Tsang,5,6 Manuel A. Rivas,7 Matti Pirinen,8 Maria Gutierrez-Arcelus,9 Kevin S. Smith,5,10 Kim R. Kukurba,5,10 Rui Zhang,10 Celeste Eng,2 Dara G. Torgerson,2 Cydney Urbanek,11 the GTEx Consortium, Jin Billy Li,10 Jose R. Rodriguez-Santana,12 Esteban G. Burchard,2,13 Max A. Seibold,11,14,15 Daniel G. MacArthur,3,4,16 Stephen B. Montgomery,5,10 Noah A. Zaitlen,2,19 and Tuuli Lappalainen17,18,19 1The Blavatnik School of Computer Science, Tel-Aviv University, Tel Aviv 69978, Israel; 2Department of Medicine, University of California San Francisco, San Francisco, California 94158, USA; 3Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts 02114, USA; 4Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, Massachusetts 02142, USA; 5Department of Pathology, Stanford University, Stanford, California 94305, USA; 6Biomedical Informatics Program, Stanford University, Stanford, California 94305, USA; 7Wellcome Trust Center for Human Genetics, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, OX3 7BN, United Kingdom; 8Institute for Molecular Medicine Finland, University of Helsinki, 00014 Helsinki, Finland; 9Department of Genetic Medicine and Development, University of Geneva, 1211 Geneva, Switzerland; -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Communication in Asthmatic Airways Inflammation and Intercellular Gene
Gene Expression Patterns of Th2 Inflammation and Intercellular Communication in Asthmatic Airways This information is current as David F. Choy, Barmak Modrek, Alexander R. Abbas, Sarah of September 28, 2021. Kummerfeld, Hilary F. Clark, Lawren C. Wu, Grazyna Fedorowicz, Zora Modrusan, John V. Fahy, Prescott G. Woodruff and Joseph R. Arron J Immunol 2011; 186:1861-1869; Prepublished online 27 December 2010; Downloaded from doi: 10.4049/jimmunol.1002568 http://www.jimmunol.org/content/186/3/1861 Supplementary http://www.jimmunol.org/content/suppl/2010/12/27/jimmunol.100256 http://www.jimmunol.org/ Material 8.DC1 References This article cites 63 articles, 10 of which you can access for free at: http://www.jimmunol.org/content/186/3/1861.full#ref-list-1 Why The JI? Submit online. by guest on September 28, 2021 • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Gene Expression Patterns of Th2 Inflammation and Intercellular Communication in Asthmatic Airways David F. -

Dynamic Interplay Between Locus-Specific DNA Methylation and Hydroxymethylation Regulates Distinct Biological Pathways in Prostate Carcinogenesis Shivani N
Kamdar et al. Clinical Epigenetics (2016) 8:32 DOI 10.1186/s13148-016-0195-4 RESEARCH Open Access Dynamic interplay between locus-specific DNA methylation and hydroxymethylation regulates distinct biological pathways in prostate carcinogenesis Shivani N. Kamdar1,2, Linh T. Ho1, Ken J. Kron1, Ruth Isserlin3, Theodorus van der Kwast1,4, Alexandre R. Zlotta5, Neil E. Fleshner6, Gary Bader3 and Bharati Bapat1,2* Abstract Background: Despite the significant global loss of DNA hydroxymethylation marks in prostate cancer tissues, the locus-specific role of hydroxymethylation in prostate tumorigenesis is unknown. We characterized hydroxymethylation and methylation marks by performing whole-genome next-generation sequencing in representative normal and prostate cancer-derived cell lines in order to determine functional pathways and key genes regulated by these epigenomic modifications in cancer. Results: Our cell line model shows disruption of hydroxymethylation distribution in cancer, with global loss and highly specific gain in promoter and CpG island regions. Significantly, we observed locus-specific retention of hydroxymethylation marks in specific intronic and intergenic regions which may play a novel role in the regulation of gene expression in critical functional pathways, such as BARD1 signaling and steroid hormone receptor signaling in cancer. We confirm a modest correlation of hydroxymethylation with expression in intragenic regions in prostate cancer, while identifying an original role for intergenic hydroxymethylation in differentially expressed regulatory pathways in cancer. We also demonstrate a successful strategy for the identification and validation of key candidate genes from differentially regulated biological pathways in prostate cancer. Conclusions: Our results indicate a distinct function for aberrant hydroxymethylation within each genomic feature in cancer, suggesting a specific and complex role for the deregulation of hydroxymethylation in tumorigenesis, similar to methylation. -

DDB1 Functions As a Linker to Recruit Receptor WD40 Proteins to CUL4− ROC1 Ubiquitin Ligases
Downloaded from genesdev.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press RESEARCH COMMUNICATION domain to bind with a conserved protein motif, the F DDB1 functions as a linker box, which, via its additional protein–protein interaction to recruit receptor WD40 modules, recruits various substrates, often phosphory- lated, to the CUL1–ROC1 catalytic core. To bring spe- proteins to CUL4–ROC1 cific substrates to CUL2- and CUL5-dependent ligases, a ubiquitin ligases heterodimeric linker complex containing elongins B and C binds simultaneously to an analogous N-terminal do- Yizhou Joseph He, Chad M. McCall, Jian Hu, main in CUL2 and CUL5 and to two similar protein 1 motifs, the VHL box and SOCS box. VHL and SOCS pro- Yaxue Zeng, and Yue Xiong teins, via their additional protein–protein interaction Lineberger Comprehensive Cancer Center, Department of modules, target various substrates differentially to the Biochemistry and Biophysics, Program in Molecular Biology CUL2–ROC1 or CUL5–ROC2 catalytic cores (Kamura et and Biotechnology, University of North Carolina, Chapel Hill, al. 1998, 2001, 2004; Stebbins et al. 1999; Zhang et al. North Carolina 27599, USA 1999). Omitting a linker, CUL3 utilizes its N-terminal domain to bind to proteins with a conserved 100-residue Cullins assemble the largest family of ubiquitin ligases protein motif known as a BTB domain, which, via addi- by binding with ROC1 and various substrate receptors. tional protein–protein interaction domains, then target CUL4 function is linked with many cellular processes, various substrates to the CUL3–ROC1 catalytic core (Fu- rukawa et al. 2003; Geyer et al. -

Chip-Seq Analysis to Explore DNA Replication Profile in Trifluridine
ANTICANCER RESEARCH 39 : 3565-3570 (2019) doi:10.21873/anticanres.13502 ChIP-seq Analysis to Explore DNA Replication Profile in Trifluridine-treated Human Colorectal Cancer Cells In Vitro TAKASHI KOBUNAI 1* , KAZUAKI MATSUOKA 2* and TEIJI TAKECHI 1 1Taiho Pharmaceutical Co., Ltd., Translational Research Laboratory, Tokyo, Japan; 2Taiho Pharmaceutical Co., Ltd., Translational Research Laboratory (Tokushima office), Tokushima, Japan Abstract. Background/Aim: Trifluridine (FTD) is a key efficacy and a tolerable safety profile for previously treated component of the novel oral antitumor drug trifluridine/ metastatic colorectal cancer (2). tipiracil that has been approved for the treatment of metastatic In basic research, FTD, the active antitumor component of colorectal cancer. In this study, a comprehensive analysis of FTD/TPI, is an antineoplastic thymidine analog (3), efficiently DNA replication profile in FTD-treated colon cancer cells was incorporated into genomic DNA in tumor cells (4-7). FTD performed. Materials and Methods: HCT-116 cells were transported into the cytoplasm of tumor cells by equilibrative exposed to BrdU or FTD and subjected to DNA immuno- nucleoside transporter 1 and 2 is phosphorylated to mono- precipitation. Immunoprecipitated DNA was sequenced; the phosphate, diphosphate, and triphosphate (FTD-TP) forms by density of aligned reads along the genome was calculated. Peak thymidine kinase 1 and nucleoside diphosphate kinase, finding, gene ontology, and motif analysis were performed using respectively. DNA polymerase α incorporates FTD-TP into MACS, GREAT, and MEME, respectively. Results: We identified DNA at sites aligned with adenine on the opposite strand (8). 6,043 and 5,080 high-confidence FTD and BrdU peaks in HCT- FTD/TPI exerts its antitumor activity in vivo predominantly 116 cells, respectively. -

Chromosome 20 Shows Linkage with DSM-IV Nicotine Dependence in Finnish Adult Smokers
Nicotine & Tobacco Research, Volume 14, Number 2 (February 2012) 153–160 Original Investigation Chromosome 20 Shows Linkage With DSM-IV Nicotine Dependence in Finnish Adult Smokers Kaisu Keskitalo-Vuokko, Ph.D.,1 Jenni Hällfors, M.Sc.,1,2 Ulla Broms, Ph.D.,1,3 Michele L. Pergadia, Ph.D.,4 Scott F. Saccone, Ph.D.,4 Anu Loukola, Ph.D.,1,3 Pamela A. F. Madden, Ph.D.,4 & Jaakko Kaprio, M.D., Ph.D.1,2,3 1 Hjelt Institute, Department of Public Health, University of Helsinki, Helsinki, Finland 2 Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland 3 National Institute for Health and Welfare (THL), Helsinki, Finland 4 Department of Psychiatry, Washington University School of Medicine, St. Louis, MO Corresponding Author: Jaakko Kaprio, M.D., Ph.D., Department of Public Health, University of Helsinki, PO Box 41 (Manner- heimintie 172), Helsinki 00014, Finland. Telephone: +358-9-191-27595; Fax: +358-9-19127570; E-mail: [email protected] Received February 5, 2011; accepted June 21, 2011 2009). Despite several gene-mapping studies, the genes underlying Abstract liability to nicotine dependence (ND) remain largely unknown. Introduction: Chromosome 20 has previously been associated Recently, Han, Gelernter, Luo, and Yang (2010) performed a with nicotine dependence (ND) and smoking cessation. Our meta-analysis of 15 genome-wide linkage scans of smoking aim was to replicate and extend these findings. behavior. Linkage signals were observed on chromosomal regions 17q24.3–q25.3, 5q33.1–q35.2, 20q13.12–32, and 22q12.3–13.32. Methods: First, a total of 759 subjects belonging to 206 Finnish The relevance of the chromosome 20 finding is highlighted families were genotyped with 18 microsatellite markers residing by the fact that CHRNA4 encoding the nicotinic acetylcholine on chromosome 20, in order to replicate previous linkage findings. -

Functional Specialization of Human Salivary Glands and Origins of Proteins Intrinsic to Human Saliva
UCSF UC San Francisco Previously Published Works Title Functional Specialization of Human Salivary Glands and Origins of Proteins Intrinsic to Human Saliva. Permalink https://escholarship.org/uc/item/95h5g8mq Journal Cell reports, 33(7) ISSN 2211-1247 Authors Saitou, Marie Gaylord, Eliza A Xu, Erica et al. Publication Date 2020-11-01 DOI 10.1016/j.celrep.2020.108402 Peer reviewed eScholarship.org Powered by the California Digital Library University of California HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Cell Rep Manuscript Author . Author manuscript; Manuscript Author available in PMC 2020 November 30. Published in final edited form as: Cell Rep. 2020 November 17; 33(7): 108402. doi:10.1016/j.celrep.2020.108402. Functional Specialization of Human Salivary Glands and Origins of Proteins Intrinsic to Human Saliva Marie Saitou1,2,3, Eliza A. Gaylord4, Erica Xu1,7, Alison J. May4, Lubov Neznanova5, Sara Nathan4, Anissa Grawe4, Jolie Chang6, William Ryan6, Stefan Ruhl5,*, Sarah M. Knox4,*, Omer Gokcumen1,8,* 1Department of Biological Sciences, University at Buffalo, The State University of New York, Buffalo, NY, U.S.A 2Section of Genetic Medicine, Department of Medicine, University of Chicago, Chicago, IL, U.S.A 3Faculty of Biosciences, Norwegian University of Life Sciences, Ås, Viken, Norway 4Program in Craniofacial Biology, Department of Cell and Tissue Biology, School of Dentistry, University of California, San Francisco, CA, U.S.A 5Department of Oral Biology, School of Dental Medicine, University at Buffalo, The State University of New York, Buffalo, NY, U.S.A 6Department of Otolaryngology, School of Medicine, University of California, San Francisco, CA, U.S.A 7Present address: Weill-Cornell Medical College, Physiology and Biophysics Department 8Lead Contact SUMMARY Salivary proteins are essential for maintaining health in the oral cavity and proximal digestive tract, and they serve as potential diagnostic markers for monitoring human health and disease. -

Development and Validation of a Serum Biomarker Panel for The
Journal of Cancer 2017, Vol. 8 2346 Ivyspring International Publisher Journal of Cancer 2017; 8(12): 2346-2355. doi: 10.7150/jca.19465 Research Paper Development and Validation of a Serum Biomarker Panel for the Detection of Esophageal Squamous Cell Carcinoma through RNA Transcriptome Sequencing Shan Xing1,2†, Xin Zheng1,2†, Li-Qiang Wei4†, Shi-Jian Song5, Dan Liu1,3, Ning Xue1,3, Xiao-Min Liu1,2, Mian-Tao Wu1,2, Qian Zhong1,3, Chu-Mei Huang6, Mu-Sheng Zeng1,3, Wan-Li Liu1,2 1. State Key Laboratory of Oncology in Southern China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China 2. Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, Guangzhou, China 3. Department of Experimental Research, Sun Yat-sen University cancer center, Guangzhou, China 4. Department of Clinical Laboratory, Shaanxi Provincial People’s Hospital, Xian, China 5. Guangdong Experimental High School, Guangzhou, China 6. Department of Laboratory Medicine, Sun Yat-sen University First Affiliated Hospital, Guangzhou, China † Xing S, Zheng X and Wei LQ contributed equally to this work Corresponding authors: Wan-Li Liu. Email: [email protected]; Phone: 86-020-87343196. Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East,Guangzhou 510060, P.R. China. Mu-Sheng Zeng; Email: [email protected] Phone: +86-020-87343192. Department of Experimental Research, Sun Yat-sen University cancer center, 651 Dongfeng Road East,Guangzhou 510060, P.R.China © Ivyspring International Publisher. This is an open access article distributed under the terms of the Creative Commons Attribution (CC BY-NC) license (https://creativecommons.org/licenses/by-nc/4.0/). -

Expression and Purification of Functional Recombinant CUL2
www.nature.com/scientificreports OPEN Expression and purifcation of functional recombinant CUL2•RBX1 from E. coli Stephanie Diaz1, Lihong Li1,2, Kankan Wang1 & Xing Liu1,2* Cullin-2 (CUL2) based cullin-RING ligases (CRL2s) comprise a family of ubiquitin E3 ligases that exist only in multi-cellular organisms and are crucial for cellular processes such as embryogenesis and viral pathogenesis. CUL2 is the scafold protein that binds one of the interchangeable substrate receptor modules, which consists of adaptor proteins and the substrate receptor protein. The VHL protein is a substrate receptor known to target hypoxia-inducible factor α (HIF1α) for ubiquitination and degradation. Because of its critical role in the ubiquitination of important cellular factors such as HIF1α, CRL2s have been investigated for their biological functions and the development of novel therapeutics against diseases. Given the importance of CRL2s in biological and biomedical research, methods that efciently produce functional CUL2 proteins will greatly facilitate studies on the mechanism and regulation of CRL2s. Here, we report two cost-efective systems for the expression and purifcation of recombinant human CUL2 from E. coli cells. The purifed CUL2 proteins were ~ 95% pure, could bind their substrate receptor modules, and were enzymatically active in transferring ubiquitin or ubiquitin-like protein to the corresponding substrate in in vitro assays. The presented methodological advancements will help advance research in CRL2 function and regulation. Protein turnover is a cellular regulatory system defned by the continuous synthesis and decomposition of specifc proteins to maintain the integrity of optimally functioning proteins 1,2. Abnormalities during protein turnover, specifcally during protein degradation, ofen result in human diseases such as cystic fbrosis and liposarcoma. -

Catenin/T-Cell Factor Signaling Identified by Gene Expression Profiling of Ovarian Endometrioid Adenocarcinomas1
[CANCER RESEARCH 63, 2913–2922, June 1, 2003] Novel Candidate Targets of -Catenin/T-cell Factor Signaling Identified by Gene Expression Profiling of Ovarian Endometrioid Adenocarcinomas1 Donald R. Schwartz,2 Rong Wu,2 Sharon L. R. Kardia, Albert M. Levin, Chiang-Ching Huang, Kerby A. Shedden, Rork Kuick, David E. Misek, Samir M. Hanash, Jeremy M. G. Taylor, Heather Reed, Neali Hendrix, Yali Zhai, Eric R. Fearon, and Kathleen R. Cho3 Comprehensive Cancer Center and Departments of Pathology [D. R. S., R. W., H. R., N. H., Y. Z., E. R. F., K. R. C.], Internal Medicine [E. R. F., K. R. C.], Pediatrics and Communicable Diseases [R. K., D. E. M., S. M. H.], and Biostatistics [C-C. H., J. M. G. T.], School of Medicine, Department of Epidemiology [S. L. R. K., A. M. L.], School of Public Health and Statistics [K. A. S.], College of Literature Science and the Arts, University of Michigan, Ann Arbor, Michigan 48109-0638 ABSTRACT specific molecular and pathobiological features [reviewed by Feeley and Wells (2) and Aunoble et al. (3)].   The activity of -catenin ( -cat), a key component of the Wnt signaling OEAs4 are characterized by frequent (16–54%) mutations of pathway, is deregulated in about 40% of ovarian endometrioid adenocar- CTNNB1, the gene encoding -cat, a critical component of the Wnt cinomas (OEAs), usually as a result of CTNNB1 gene mutations. The signaling pathway (4–9). Wnts are a highly conserved family of function of -cat in neoplastic transformation is dependent on T-cell factor (TCF) transcription factors, but specific genes activated by the secreted growth factors that bind members of the frizzled family of interaction of -cat with TCFs in OEAs and other cancers with Wnt transmembrane receptors and, through downstream signaling, modu- pathway defects are largely unclear. -
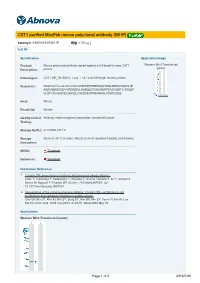
CST1 Purified Maxpab Mouse Polyclonal Antibody (B01P)
CST1 purified MaxPab mouse polyclonal antibody (B01P) Catalog # : H00001469-B01P 規格 : [ 50 ug ] List All Specification Application Image Product Mouse polyclonal antibody raised against a full-length human CST1 Western Blot (Transfected lysate) Description: protein. Immunogen: CST1 (NP_001889.2, 1 a.a. ~ 141 a.a) full-length human protein. Sequence: MAQYLSTLLLLLATLAVALAWSPKEEDRIIPGGIYNADLNDEWVQRALHF AISEYNKATKDDYYRRPLRVLRARQQTVGGVNYFFDVEVGRTICTKSQP NLDTCAFHEQPELQKKQLCSFEIYEVPWENRRSLVKSRCQES enlarge Host: Mouse Reactivity: Human Quality Control Antibody reactive against mammalian transfected lysate. Testing: Storage Buffer: In 1x PBS, pH 7.4 Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Instruction: MSDS: Download Datasheet: Download Publication Reference 1. Cystatin SN Upregulation in Patients with Seasonal Allergic Rhinitis. Imoto Y, Tokunaga T, Matsumoto Y, Hamada Y, Ono M, Yamada T, Ito Y, Arinami T, Okano M, Noguchi E, Fujieda SPLoS One. 2013;8(8):e67057. doi: 10.1371/journal.pone.0067057. 2. Upregulation of the cysteine protease inhibitor, Cystatin SN, contributes to cell proliferation and cathepsin inhibition in gastric cancer. Choi EH, Kim JT, Kim JH, Kim SY, Song EY, Kim JW, Kim SY, Yeom YI, Kim IH, Lee HG.Clin Chim Acta. 2009 Aug;406(1-2):45-51. Epub 2009 May 19. Applications Western Blot (Transfected lysate) Page 1 of 2 2016/5/20 Western Blot analysis of CST1 expression in transfected 293T cell line (H00001469-T01) by CST1 MaxPab polyclonal antibody. Lane 1: CST1 transfected lysate(15.51 KDa). Lane 2: Non-transfected lysate. Protocol Download Gene Information Entrez GeneID: 1469 GeneBank NM_001898.2 Accession#: Protein NP_001889.2 Accession#: Gene Name: CST1 Gene Alias: - Gene cystatin SN Description: Omim ID: 123855 Gene Ontology: Hyperlink Gene Summary: The cystatin superfamily encompasses proteins that contain multiple cystatin-like sequences.