Dynamic Interplay Between Locus-Specific DNA Methylation and Hydroxymethylation Regulates Distinct Biological Pathways in Prostate Carcinogenesis Shivani N
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

RET Gene Fusions in Malignancies of the Thyroid and Other Tissues
G C A T T A C G G C A T genes Review RET Gene Fusions in Malignancies of the Thyroid and Other Tissues Massimo Santoro 1,*, Marialuisa Moccia 1, Giorgia Federico 1 and Francesca Carlomagno 1,2 1 Department of Molecular Medicine and Medical Biotechnology, University of Naples “Federico II”, 80131 Naples, Italy; [email protected] (M.M.); [email protected] (G.F.); [email protected] (F.C.) 2 Institute of Endocrinology and Experimental Oncology of the CNR, 80131 Naples, Italy * Correspondence: [email protected] Received: 10 March 2020; Accepted: 12 April 2020; Published: 15 April 2020 Abstract: Following the identification of the BCR-ABL1 (Breakpoint Cluster Region-ABelson murine Leukemia) fusion in chronic myelogenous leukemia, gene fusions generating chimeric oncoproteins have been recognized as common genomic structural variations in human malignancies. This is, in particular, a frequent mechanism in the oncogenic conversion of protein kinases. Gene fusion was the first mechanism identified for the oncogenic activation of the receptor tyrosine kinase RET (REarranged during Transfection), initially discovered in papillary thyroid carcinoma (PTC). More recently, the advent of highly sensitive massive parallel (next generation sequencing, NGS) sequencing of tumor DNA or cell-free (cfDNA) circulating tumor DNA, allowed for the detection of RET fusions in many other solid and hematopoietic malignancies. This review summarizes the role of RET fusions in the pathogenesis of human cancer. Keywords: kinase; tyrosine kinase inhibitor; targeted therapy; thyroid cancer 1. The RET Receptor RET (REarranged during Transfection) was initially isolated as a rearranged oncoprotein upon the transfection of a human lymphoma DNA [1]. -

Genome-Wide Association Study Identifies NRG1 As a Susceptibility Locus for Hirschsprung’S Disease
Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease Maria-Merce` Garcia-Barceloa,b,1, Clara Sze-man Tangc,1, Elly Sau-wai Ngana,b, Vincent Chi-hang Luia,b, Yan Chena, Man-ting Soa, Thomas Yuk-yu Leona, Xiao-ping Miaoa,d, Cathy Ka-yee Shuma, Feng-qin Liua, Ming-yiu Yeungc, Zhen-wei Yuane, Wei-hong Guof, Lei Liuc, Xiao-bing Sung, Liu-ming Huangh, Jin-fa Toui, You-qiang Songj, Danny Chanj, Kenneth M. C. Cheungk, Kenneth Kak-yuen Wonga, Stacey S. Chernyc,l,2, Pak-chung Shamb,c,2, and Paul Kwong-hang Tama,b,2 Departments of aSurgery, cDepartment of Psychiatry, lGenome Research Centre, bCentre for Reproduction, Development, and Growth, Departments of jBiochemistry and kOrthopaedics and Traumatology of the Li Ka Shing Faculty of Medicine, University of Hong Kong; eDepartment of Paediatric Surgery, Shengjing Hospital, China Medical University, Shenyang, China; fDepartment of Surgery, Beijing Children’s Hospital, Beijing, China; dDepartment of Surgery, Shenzhen Children’s Hospital, Shenzhen, China; gDepartment of Pediatric Surgery, Shandong Medical University, Shandong, China; hDepartment of Surgery, Beijing University, Beijing, China; and iDepartment of Surgery, Zhejiang Children’s Hospital, Zhejiang, China Communicated by Lap-Chee Tsui, University of Hong Kong, Hong Kong SAR, People’s Republic of China, September 30, 2008 (received for review June 24, 2008) Hirschsprung’s disease (HSCR), or aganglionic megacolon, is a the length of the aganglionic segment in the proband and the congenital disorder characterized by the absence of enteric ganglia gender of the sibling. in variable portions of the distal intestine. -

DDB1 Functions As a Linker to Recruit Receptor WD40 Proteins to CUL4− ROC1 Ubiquitin Ligases
Downloaded from genesdev.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press RESEARCH COMMUNICATION domain to bind with a conserved protein motif, the F DDB1 functions as a linker box, which, via its additional protein–protein interaction to recruit receptor WD40 modules, recruits various substrates, often phosphory- lated, to the CUL1–ROC1 catalytic core. To bring spe- proteins to CUL4–ROC1 cific substrates to CUL2- and CUL5-dependent ligases, a ubiquitin ligases heterodimeric linker complex containing elongins B and C binds simultaneously to an analogous N-terminal do- Yizhou Joseph He, Chad M. McCall, Jian Hu, main in CUL2 and CUL5 and to two similar protein 1 motifs, the VHL box and SOCS box. VHL and SOCS pro- Yaxue Zeng, and Yue Xiong teins, via their additional protein–protein interaction Lineberger Comprehensive Cancer Center, Department of modules, target various substrates differentially to the Biochemistry and Biophysics, Program in Molecular Biology CUL2–ROC1 or CUL5–ROC2 catalytic cores (Kamura et and Biotechnology, University of North Carolina, Chapel Hill, al. 1998, 2001, 2004; Stebbins et al. 1999; Zhang et al. North Carolina 27599, USA 1999). Omitting a linker, CUL3 utilizes its N-terminal domain to bind to proteins with a conserved 100-residue Cullins assemble the largest family of ubiquitin ligases protein motif known as a BTB domain, which, via addi- by binding with ROC1 and various substrate receptors. tional protein–protein interaction domains, then target CUL4 function is linked with many cellular processes, various substrates to the CUL3–ROC1 catalytic core (Fu- rukawa et al. 2003; Geyer et al. -

Mai Muudatuntuu Ti on Man Mini
MAIMUUDATUNTUU US009809854B2 TI ON MAN MINI (12 ) United States Patent ( 10 ) Patent No. : US 9 ,809 ,854 B2 Crow et al. (45 ) Date of Patent : Nov . 7 , 2017 Whitehead et al. (2005 ) Variation in tissue - specific gene expression ( 54 ) BIOMARKERS FOR DISEASE ACTIVITY among natural populations. Genome Biology, 6 :R13 . * AND CLINICAL MANIFESTATIONS Villanueva et al. ( 2011 ) Netting Neutrophils Induce Endothelial SYSTEMIC LUPUS ERYTHEMATOSUS Damage , Infiltrate Tissues, and Expose Immunostimulatory Mol ecules in Systemic Lupus Erythematosus . The Journal of Immunol @(71 ) Applicant: NEW YORK SOCIETY FOR THE ogy , 187 : 538 - 552 . * RUPTURED AND CRIPPLED Bijl et al. (2001 ) Fas expression on peripheral blood lymphocytes in MAINTAINING THE HOSPITAL , systemic lupus erythematosus ( SLE ) : relation to lymphocyte acti vation and disease activity . Lupus, 10 :866 - 872 . * New York , NY (US ) Crow et al . (2003 ) Microarray analysis of gene expression in lupus. Arthritis Research and Therapy , 5 :279 - 287 . * @(72 ) Inventors : Mary K . Crow , New York , NY (US ) ; Baechler et al . ( 2003 ) Interferon - inducible gene expression signa Mikhail Olferiev , Mount Kisco , NY ture in peripheral blood cells of patients with severe lupus . PNAS , (US ) 100 ( 5 ) : 2610 - 2615. * GeneCards database entry for IFIT3 ( obtained from < http : / /www . ( 73 ) Assignee : NEW YORK SOCIETY FOR THE genecards. org /cgi - bin / carddisp .pl ? gene = IFIT3 > on May 26 , 2016 , RUPTURED AND CRIPPLED 15 pages ) . * Navarra et al. (2011 ) Efficacy and safety of belimumab in patients MAINTAINING THE HOSPITAL with active systemic lupus erythematosus : a randomised , placebo FOR SPECIAL SURGERY , New controlled , phase 3 trial . The Lancet , 377 :721 - 731. * York , NY (US ) Abramson et al . ( 1983 ) Arthritis Rheum . -

Expression and Purification of Functional Recombinant CUL2
www.nature.com/scientificreports OPEN Expression and purifcation of functional recombinant CUL2•RBX1 from E. coli Stephanie Diaz1, Lihong Li1,2, Kankan Wang1 & Xing Liu1,2* Cullin-2 (CUL2) based cullin-RING ligases (CRL2s) comprise a family of ubiquitin E3 ligases that exist only in multi-cellular organisms and are crucial for cellular processes such as embryogenesis and viral pathogenesis. CUL2 is the scafold protein that binds one of the interchangeable substrate receptor modules, which consists of adaptor proteins and the substrate receptor protein. The VHL protein is a substrate receptor known to target hypoxia-inducible factor α (HIF1α) for ubiquitination and degradation. Because of its critical role in the ubiquitination of important cellular factors such as HIF1α, CRL2s have been investigated for their biological functions and the development of novel therapeutics against diseases. Given the importance of CRL2s in biological and biomedical research, methods that efciently produce functional CUL2 proteins will greatly facilitate studies on the mechanism and regulation of CRL2s. Here, we report two cost-efective systems for the expression and purifcation of recombinant human CUL2 from E. coli cells. The purifed CUL2 proteins were ~ 95% pure, could bind their substrate receptor modules, and were enzymatically active in transferring ubiquitin or ubiquitin-like protein to the corresponding substrate in in vitro assays. The presented methodological advancements will help advance research in CRL2 function and regulation. Protein turnover is a cellular regulatory system defned by the continuous synthesis and decomposition of specifc proteins to maintain the integrity of optimally functioning proteins 1,2. Abnormalities during protein turnover, specifcally during protein degradation, ofen result in human diseases such as cystic fbrosis and liposarcoma. -

The Structure and Regulation of Cullin 2 Based E3 Ubiquitin Ligases and Their Biological Functions Weijia Cai* and Haifeng Yang*
Cai and Yang Cell Div (2016) 11:7 DOI 10.1186/s13008-016-0020-7 Cell Division REVIEW Open Access The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions Weijia Cai* and Haifeng Yang* Abstract Background: Cullin-RING E3 ubiquitin ligase complexes play a central role in targeting cellular proteins for ubiquit- ination-dependent protein turnover through 26S proteasome. Cullin-2 is a member of the Cullin family, and it serves as a scaffold protein for Elongin B and C, Rbx1 and various substrate recognition receptors to form E3 ubiquitin ligases. Main body of the abstract: First, the composition, structure and the regulation of Cullin-2 based E3 ubiquitin ligases were introduced. Then the targets, the biological functions of complexes that use VHL, Lrr-1, Fem1b, Prame, Zyg-11, BAF250, Rack1 as substrate targeting subunits were described, and their involvement in diseases was discussed. A small molecule inhibitor of Cullins as a potential anti-cancer drug was introduced. Furthermore, proteins with VHL box that might bind to Cullin-2 were described. Finally, how different viral proteins form E3 ubiquitin ligase complexes with Cullin-2 to counter host viral defense were explained. Conclusions: Cullin-2 based E3 ubiquitin ligases, using many different substrate recognition receptors, recognize a number of substrates and regulate their protein stability. These complexes play critical roles in biological processes and diseases such as cancer, germline differentiation and viral defense. Through the better understanding of their biology, we can devise and develop new therapeutic strategies to treat cancers, inherited diseases and viral infections. -

Modeling Genomic Diversity and Tumor Dependency in Malignant Melanoma
Research Article Modeling Genomic Diversity and Tumor Dependency in Malignant Melanoma William M. Lin,1,3,5 Alissa C. Baker,1,3 Rameen Beroukhim,1,3,5 Wendy Winckler,1,3,5 Whei Feng,1,3,5 Jennifer M. Marmion,7 Elisabeth Laine,8 Heidi Greulich,1,3,5 Hsiuyi Tseng,1,3 Casey Gates,5 F. Stephen Hodi,1 Glenn Dranoff,1 William R. Sellers,1,6 Roman K. Thomas,9,10 Matthew Meyerson,1,3,4,5 Todd R. Golub,2,3,5 Reinhard Dummer,8 Meenhard Herlyn,7 Gad Getz,3,5 and Levi A. Garraway1,3,5 Departments of 1Medical Oncology and 2Pediatric Oncology and 3Center for Cancer Genome Discovery, Dana-Farber Cancer Institute, Harvard Medical School; 4Department of Pathology, Harvard Medical School, Boston, Massachusetts; 5The Broad Institute of M.I.T. and Harvard; 6Novartis Institutes for Biomedical Research, Cambridge, Massachusetts; 7Cancer Biology Division, Wistar Institute, Philadelphia, Pennsylvania; 8Department of Dermatology, University of Zurich Hospital, Zu¨rich, Switzerland; 9Max Planck Institute for Neurological Research with Klaus Joachim Zulch Laboratories of the Max Planck Society and the Medical Faculty of the University of Cologne; and 10Center for Integrated Oncology and Department I for Internal Medicine, University of Cologne, Cologne, Germany Abstract tumorigenesis have been derived from functional studies involving The classification of human tumors based on molecular cultured human cancer cells (e.g., established cell lines, short-term cultures, etc.). Despite their limitations, cancer cell line collections criteria offers tremendous clinical potential; however, dis- cerning critical and ‘‘druggable’’ effectors on a large scale will whose genetic alterations reflect their primary tumor counterparts also require robust experimental models reflective of tumor should provide malleable proxies that facilitate mechanistic genomic diversity. -

A 25-Gene Classifier Predicts Overall Survival in Resectable Pancreatic Cancer David J
Birnbaum et al. BMC Medicine (2017) 15:170 DOI 10.1186/s12916-017-0936-z RESEARCHARTICLE Open Access A 25-gene classifier predicts overall survival in resectable pancreatic cancer David J. Birnbaum1,2,3, Pascal Finetti1, Alexia Lopresti1, Marine Gilabert4, Flora Poizat5, Jean-Luc Raoul4, Jean-Robert Delpero3,6, Vincent Moutardier2,3, Daniel Birnbaum1, Emilie Mamessier1† and François Bertucci1,3,4,7*† Abstract Background: Pancreatic carcinoma is one of the most lethal human cancers. In patients with resectable tumors, surgery followed by adjuvant chemotherapy is the only curative treatment. However, the 5-year survival is 20%. Because of a strong metastatic propensity, neoadjuvant chemotherapy is being tested in randomized clinical trials. In this context, improving the selection of patients for immediate surgery or neoadjuvant chemotherapy is crucial, and high-throughput molecular analyses may help; the present study aims to address this. Methods: Clinicopathological and gene expression data of 695 pancreatic carcinoma samples were collected from nine datasets and supervised analysis was applied to search for a gene expression signature predictive for overall survival (OS) in the 601 informative operated patients. The signature was identified in a learning set of patients and tested for its robustness in a large independent validation set. Results: Supervised analysis identified 1400 genes differentially expressed between two selected patient groups in the learning set, namely 17 long-term survivors (LTS; ≥ 36 months after surgery) and 22 short-term survivors (STS; dead of disease between 2 and 6 months after surgery). From these, a 25-gene prognostic classifier was developed, which identified two classes (“STS-like” and “LTS-like”) in the independent validation set (n = 562), with a 25% (95% CI 18–33) and 48% (95% CI 42–54) 2-year OS (P = 4.33 × 10–9), respectively. -

Short Article CAND1 Binds to Unneddylated CUL1 and Regulates
Molecular Cell, Vol. 10, 1519–1526, December, 2002, Copyright 2002 by Cell Press CAND1 Binds to Unneddylated CUL1 Short Article and Regulates the Formation of SCF Ubiquitin E3 Ligase Complex Jianyu Zheng,1 Xiaoming Yang,1 Despite the importance of cullins in controlling many Jennifer M. Harrell,1 Sophia Ryzhikov,1 essential biological processes, the mechanism that reg- Eun-Hee Shim,1 Karin Lykke-Andersen,2 ulates the cullin-containing ubiquitin E3 ligases remains Ning Wei,2 Hong Sun,1 Ryuji Kobayashi,3 unclear. In SCF, the F box proteins are short-lived pro- and Hui Zhang1,4 teins that undergo CUL1/SKP1-dependent degradation 1Department of Genetics (Wirbelauer et al., 2000; Zhou and Howley, 1998). Dele- Yale University School of Medicine tion of the F box region abolishes the binding of F box 333 Cedar Street proteins to SKP1 and CUL1, and consequently increases 2 Department of Molecular, Cellular, the stability of F box proteins. This substrate-indepen- and Developmental Biology dent proteolysis of F box proteins is likely the result of Yale University autoubiquitination by the ubiquitin E2 and E1 enzymes New Haven, Connecticut 06520 through a CUL1/SKP1-dependent mechanism. 3 Cold Spring Harbor Laboratory The carboxy-terminal ends of cullins are often covalently Cold Spring Harbor, New York 11724 modified by a ubiquitin-like protein, NEDD8/RUB1, and this modification appears to associate with active E3 li- gases (Hochstrasser, 2000). Like ubiquitin modification, Summary neddylation requires E1 (APP-BP1 and UBA3)-activating and E2 (UBC12)-conjugating enzymes (Hochstrasser, The SCF ubiquitin E3 ligase regulates ubiquitin-depen- 2000). -

Downloaded from the Broad Institute (
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by eScholarship - University of California UCLA UCLA Previously Published Works Title A genome-wide association study identifies potential susceptibility loci for Hirschsprung disease. Permalink https://escholarship.org/uc/item/9076t8wt Journal PloS one, 9(10) ISSN 1932-6203 Authors Kim, Jeong-Hyun Cheong, Hyun Sub Sul, Jae Hoon et al. Publication Date 2014 DOI 10.1371/journal.pone.0110292 Peer reviewed eScholarship.org Powered by the California Digital Library University of California A Genome-Wide Association Study Identifies Potential Susceptibility Loci for Hirschsprung Disease Jeong-Hyun Kim1,2, Hyun Sub Cheong3, Jae Hoon Sul4, Jeong-Meen Seo5, Dae-Yeon Kim6, Jung-Tak Oh7, Kwi-Won Park8, Hyun-Young Kim8, Soo-Min Jung5, Kyuwhan Jung9, Min Jeng Cho10, Joon Seol Bae11, Hyoung Doo Shin1,2,3* 1 Research Institute for Basic Science, Sogang University, Seoul, Republic of Korea, 2 Department of Life Science, Sogang University, Seoul, Republic of Korea, 3 Department of Genetic Epidemiology, SNP Genetics, Inc., Seoul, Republic of Korea, 4 Department of Computer Science, University of California Los Angeles, Los Angeles, California, United States of America, 5 Division of Pediatric Surgery, Department of Surgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Republic of Korea, 6 Department of Pediatric Surgery, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea, 7 Department of Pediatric Surgery, -
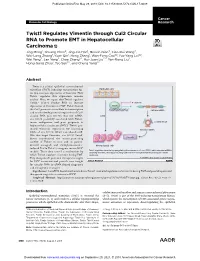
Twist1 Regulates Vimentin Through Cul2 Circular RNA to Promote EMT
Published OnlineFirst May 29, 2018; DOI: 10.1158/0008-5472.CAN-17-3009 Cancer Molecular Cell Biology Research Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma Jing Meng1, Shuang Chen2, Jing-Xia Han1, Baoxin Qian3, Xiao-Rui Wang4, Wei-Long Zhong1, Yuan Qin1, Heng Zhang1, Wan-Feng Gao1,2, Yue-Yang Lei1,2, Wei Yang2, Lan Yang2, Chao Zhang1,2, Hui-Juan Liu2,4, Yan-Rong Liu2, Hong-Gang Zhou1, Tao Sun1,2, and Cheng Yang1,2 Abstract Twist is a critical epithelial–mesenchymal transition (EMT)–inducing transcription fac- Epithelial cells tor that increases expression of vimentin. How Twist1 regulates this expression remains unclear. Here, we report that Twist1 regulates Cullin2 (Cul2) circular RNA to increase E-cadherin expressionofvimentininEMT.Twist1bound Twist1 theCul2promotertoactivateitstranscription Cul2 pre-mRNA and to selectively promote expression of Cul2 circular RNA (circ-10720), but not mRNA. EMT circ-10720 positively correlated with Twist1, tumor malignance, and poor prognosis in circRNA-10720 hepatocellular carcinoma (HCC). Twist1 pro- moted vimentin expression by increasing Vimentin miRNA levels of circ-10720, which can absorb miR- NAs that target vimentin. circ-10720 knock- down counteracted the tumor-promoting Vimentin activity of Twist1 in vitro and in patient- derived xenograft and diethylnitrosamine- Mesenchymal cells induced TetOn-Twist1 transgenic mouse HCC Twist1 regulates vimentin by upregulating the expression of circ-10720, which absorbs miRNAs models. These data unveil a mechanism by targeting vimentin, thereby promoting epithelial-mesenchymal transition in hepatocellular which Twist1 regulates vimentin during EMT. carcinoma. They also provide potential therapeutic targets © 2018 American Association for Cancer Research for HCC treatment and provide new insight forcircularRNA(circRNA)-baseddiagnostic and therapeutic strategies.