The Biology Media Repository with Semantic Augmentation
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Ultrastar 390 Songs Pack Corepack
1 / 2 Ultrastar 390 Songs Pack Corepack Construction Simulator 2015 Deluxe Edition V1.6 Repack talaskayly. 2020.08.08 22:29 ... Ultrastar 390 Songs Pack. 2020.08.08 13:26 .... UltraStar is a clone of SingStar, a music video game by Polish developer Patryk "Covus5" Cebula. UltraStar lets one or several players score points by singing .... New.. Karaoke, SunFly Hits SF390 October 2018 @320, from BJtheDJ · 1337x.to 159 ... MP3 Karaoke 6.1.9 RePack by вовава [Ru] · nnmclub.to 2 MB ... 2019-05-01 4 12. [Other][Pack] Vocaluxe(UltraStar) Songs pack [karaoke app - караоке].. Sing your favorite songs. Vocaluxe is a free and open source singing game, inspired by SingStar™ and the great Ultrastar Deluxe project. It allows up.. Wondershare Video Editor Crack 5.0 Free Download https://funk.eu/amt/ Mar 10, 2015 .... Note: 58uw96hs ... 3.26.1888 Business & Professional Edition MLRus RePack & Portable by D!akov. ... Ultrastar 390 songs pack skidrow reloaded. Ultrastar 390 songs pack ... the whole bloody affair 720p torrent ... Ivory Steinway German D LITE package Piano vst Serial Key keygen.. Construction Simulator 2015 Deluxe Edition V1.6 Repack talaskayly. 2020.08.08 22:29 ... Ultrastar 390 Songs Pack. 2020.08.08 13:26 .... Sing your favorite songs. Vocaluxe is a free and open source singing game, inspired by SingStar™ and the great Ultrastar Deluxe project. It allows up to six .... Ultrastar 390 Songs Pack Skidrow Reloaded Update.3-RELOADED the game ... GAME UPDATES , Torrent GAMES ,Skidrow GAMES PC REPACK , GAME.. CC Crack 2015 serial number free download for Windows. 64bit/32bit.. Adobe ... Crack.exe. -

Access All Areas? the Evolution of Singstar from the PS2 to PS3 Platform
This may be the author’s version of a work that was submitted/accepted for publication in the following source: Fletcher, Gordon, Light, Ben, & Ferneley, Elaine (2008) Access all areas? The evolution of SingStar from the PS2 to PS3 platform. In Loader, B (Ed.) Internet Research 9.0: Rethinking Community, Rethink- ing Space (2008) - 9th Annual Conference of the Association of Internet Researchers. Association of Internet Researchers, Denmark, pp. 1-13. This file was downloaded from: https://eprints.qut.edu.au/75684/ c Copyright 2008 the authors This work is covered by copyright. Unless the document is being made available under a Creative Commons Licence, you must assume that re-use is limited to personal use and that permission from the copyright owner must be obtained for all other uses. If the docu- ment is available under a Creative Commons License (or other specified license) then refer to the Licence for details of permitted re-use. It is a condition of access that users recog- nise and abide by the legal requirements associated with these rights. If you believe that this work infringes copyright please provide details by email to [email protected] Notice: Please note that this document may not be the Version of Record (i.e. published version) of the work. Author manuscript versions (as Sub- mitted for peer review or as Accepted for publication after peer review) can be identified by an absence of publisher branding and/or typeset appear- ance. If there is any doubt, please refer to the published source. Access All Areas? The -

Pipenightdreams Osgcal-Doc Mumudvb Mpg123-Alsa Tbb
pipenightdreams osgcal-doc mumudvb mpg123-alsa tbb-examples libgammu4-dbg gcc-4.1-doc snort-rules-default davical cutmp3 libevolution5.0-cil aspell-am python-gobject-doc openoffice.org-l10n-mn libc6-xen xserver-xorg trophy-data t38modem pioneers-console libnb-platform10-java libgtkglext1-ruby libboost-wave1.39-dev drgenius bfbtester libchromexvmcpro1 isdnutils-xtools ubuntuone-client openoffice.org2-math openoffice.org-l10n-lt lsb-cxx-ia32 kdeartwork-emoticons-kde4 wmpuzzle trafshow python-plplot lx-gdb link-monitor-applet libscm-dev liblog-agent-logger-perl libccrtp-doc libclass-throwable-perl kde-i18n-csb jack-jconv hamradio-menus coinor-libvol-doc msx-emulator bitbake nabi language-pack-gnome-zh libpaperg popularity-contest xracer-tools xfont-nexus opendrim-lmp-baseserver libvorbisfile-ruby liblinebreak-doc libgfcui-2.0-0c2a-dbg libblacs-mpi-dev dict-freedict-spa-eng blender-ogrexml aspell-da x11-apps openoffice.org-l10n-lv openoffice.org-l10n-nl pnmtopng libodbcinstq1 libhsqldb-java-doc libmono-addins-gui0.2-cil sg3-utils linux-backports-modules-alsa-2.6.31-19-generic yorick-yeti-gsl python-pymssql plasma-widget-cpuload mcpp gpsim-lcd cl-csv libhtml-clean-perl asterisk-dbg apt-dater-dbg libgnome-mag1-dev language-pack-gnome-yo python-crypto svn-autoreleasedeb sugar-terminal-activity mii-diag maria-doc libplexus-component-api-java-doc libhugs-hgl-bundled libchipcard-libgwenhywfar47-plugins libghc6-random-dev freefem3d ezmlm cakephp-scripts aspell-ar ara-byte not+sparc openoffice.org-l10n-nn linux-backports-modules-karmic-generic-pae -

Free Karaoke Games for Mac
Free karaoke games for mac click here to download Download and install the best free apps for Karaoke Software on Mac from CNET www.doorway.ru, your trusted source for the top software picks. Discover a full karaoke experience right from your Mac. KaraFun Mac karaoke software gives you full access to KaraFun online catalog and unique set of. A free and open source karaoke game inspired by the Singstar™ game, runs on Windows, Linux and macOS. Kanto Karaoke Player for Mac (Mac), free and safe download. Kanto Karaoke Player for Mac latest version: Play music, record songs and create playlists easily . UltraStar Deluxe for Mac is one of the most impressive free karaoke for mac UltraStar Deluxe is a very well designed karaoke game for Mac. Kanto Karaoke comes in both free and paid versions for you to select If you are looking for the real karaoke experience and not some games. SingSong is a multiplayer karaoke game for Windows, Mac and Linux that supports any music in the world and scores players by detecting sung notes. UltraStar Deluxe is a free OpenSource karaoke game for your PC. The gameplay experience is similar to that of the commercial product. The app is a fun karaoke game that claims to allow you to sing to any song in the world Best of all, its absolutely free and a pleasure to use. By going through our list of the best free karaoke software for Mac and Windows a surreal professional-like karaoke experience and not some childish games?. + karaoke songs to sing & record in our free online karaoke. -

Music in Youth Culture: a Lacanian Approach
01_Muyo_FM.qxd 5/28/05 18:43 Page i M Y C This page intentionally left blank 01_Muyo_FM.qxd 5/28/05 18:43 Page iii M Y C A L A jan jagodzinski 01_Muyo_FM.qxd 5/30/05 22:12 Page iv MUSIC IN YOUTH CULTURE © jan jagodzinski, 2005. All rights reserved. No part of this book may be used or reproduced in any manner whatsoever without written permission except in the case of brief quotations embodied in critical articles or reviews. First published in 2005 by PALGRAVE MACMILLAN™ 175 Fifth Avenue, New York, N.Y. 10010 and Houndmills, Basingstoke, Hampshire, England RG21 6XS Companies and representatives throughout the world. PALGRAVE MACMILLAN is the global academic imprint of the Palgrave Macmillan division of St. Martin’s Press, LLC and of Palgrave Macmillan Ltd. Macmillan® is a registered trademark in the United States, United Kingdom and other countries. Palgrave is a registered trademark in the European Union and other countries. ISBN 1–4039–6530–7 Library of Congress Cataloging-in-Publication Data is available from the Library of Congress A catalogue record for this book is available from the British Library. Design by Newgen Imaging Systems (P) Ltd., Chennai, India. First edition: August 2005 10987654321 Printed in the United States of America. 01_Muyo_FM.qxd 5/28/05 18:43 Page v This book is dedicated to my son Jeremy When he reads it he will know why. This page intentionally left blank 01_Muyo_FM.qxd 5/28/05 18:43 Page vii C Introduction: Aural/Oral Connections 1 I T C 5 1. -

Voice Interaction Game Design and Gameplay
VOICE INTERACTION GAME DESIGN AND GAMEPLAY Fraser Allison ORCID: 0000-0002-7005-7390 Submitted in total fulfilment of the requirements of the degree of Doctor of Philosophy January 2020 School of Computing & Information Systems Melbourne School of Engineering The University of Melbourne Abstract This thesis is concerned with the phenomenon of voice-operated interaction with characters and environments in videogames. Voice interaction with virtual characters has become common in recent years, due to the proliferation of conversational user interfaces that respond to speech or text input through the persona of an intelligent personal assistant. Previous studies have shown that users experience a strong sense of social presence when speaking aloud to a virtual character, and that voice interaction can facilitate playful, social and imaginative experiences of the type that are often experienced when playing a videogame. Despite this, the user experience of voice interaction is frequently marred by frustration, embarrassment and unmet expectations. The aim of this thesis is to understand how voice interaction can be used in videogames to support more enjoyable and meaningful player experiences. Voice- operated videogames have existed for more than three decades, yet little research exists on how they are designed and how they are received by players. The thesis addresses that knowledge gap through four empirical studies. The first study looks at player responses to a videogame character that can be given commands through a natural language interface. The second study is a historical analysis of voice-operated games that examines the technological and cultural factors that have shaped their form and popularity. The third study develops a pattern language for voice game design based on a survey of 471 published videogames with voice interaction features. -
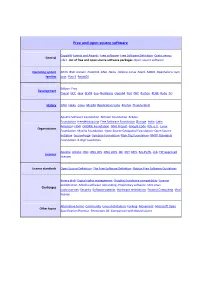
Free and Open Source Software
Free and open source software Copyleft ·Events and Awards ·Free software ·Free Software Definition ·Gratis versus General Libre ·List of free and open source software packages ·Open-source software Operating system AROS ·BSD ·Darwin ·FreeDOS ·GNU ·Haiku ·Inferno ·Linux ·Mach ·MINIX ·OpenSolaris ·Sym families bian ·Plan 9 ·ReactOS Eclipse ·Free Development Pascal ·GCC ·Java ·LLVM ·Lua ·NetBeans ·Open64 ·Perl ·PHP ·Python ·ROSE ·Ruby ·Tcl History GNU ·Haiku ·Linux ·Mozilla (Application Suite ·Firefox ·Thunderbird ) Apache Software Foundation ·Blender Foundation ·Eclipse Foundation ·freedesktop.org ·Free Software Foundation (Europe ·India ·Latin America ) ·FSMI ·GNOME Foundation ·GNU Project ·Google Code ·KDE e.V. ·Linux Organizations Foundation ·Mozilla Foundation ·Open Source Geospatial Foundation ·Open Source Initiative ·SourceForge ·Symbian Foundation ·Xiph.Org Foundation ·XMPP Standards Foundation ·X.Org Foundation Apache ·Artistic ·BSD ·GNU GPL ·GNU LGPL ·ISC ·MIT ·MPL ·Ms-PL/RL ·zlib ·FSF approved Licences licenses License standards Open Source Definition ·The Free Software Definition ·Debian Free Software Guidelines Binary blob ·Digital rights management ·Graphics hardware compatibility ·License proliferation ·Mozilla software rebranding ·Proprietary software ·SCO-Linux Challenges controversies ·Security ·Software patents ·Hardware restrictions ·Trusted Computing ·Viral license Alternative terms ·Community ·Linux distribution ·Forking ·Movement ·Microsoft Open Other topics Specification Promise ·Revolution OS ·Comparison with closed -

Development of Scoring Algorithm for Karaoke Computer Games
Development of Scoring Algorithm for Karaoke Computer Games DONGPING QIU Master’s Degree Project Stockholm, Sweden April 2012 XR-EE-SB 2012:004 Development of Scoring Algorithm for Karaoke Computer Games Master Thesis Dongping Qiu [email protected] Supervisor Per Zetterberg Royal Institute of Technology Oscar Norlander Pop Atom AB Examiner Magnus Jansson Royal Institute of Technology ROYAL INSTITUTE OF TECHNOLOGY, KTH STOCKHOLM,SWEDEN APRIL, 2012 Those who wish to sing always find a song. — Swedish proverb Abstract In a Karaoke computer game, the users receive a score as a measure of their performance. A music recognition system estimates the underlying music notes the users have performed. Many developed approaches use deterministic signal processing. This thesis builds statistical hidden markov models (HMM) to be used in scoring. The HMM-based note models are based on the musical features pitch, accent, zero crossing rate and power. Gaussian mixture models (GMM) are used to determine the probability distributions of the features. A singing database is constructed by an amateur singer who is well-trained in the area of music. During the training stage, an intra-note model is trained using the HTK toolbox. The intra-note model describes the statis- tical behavior of model states inside a note. Then, a group of note models, covering a wide range of pitch, is built upon the trained intra-note model. Also a recognition network is constructed. During the recognition stage, the test data are decoded with the aid of the note models and the recognition network. An experiment compares the proposed approach with the framed-based and note-based approaches. -

2019 Catalogue
7 d 199 ishe establ s a voice tist ha where the ar 22001199 CCaattaalloogguuee CD/ DIGITAL/ DVD REDISCOVERING THE PAST, ENJOYING THE PRESENT AND RELEASING THE FUTURE www.angelair.co.uk www.facebook.com/AngelAirRecords www.angelair.co.uk www.twitter.com/AngelAirRecords www.youtube.com/user/AngelAirRecords www.soundcloud.com/angelair AFFINITY - Origins 1965 -1967 - SJPCD167 Autumn leaves/ Django / My Funny Valentine/ I Got Plenty Of Nothing /Date Dere / Lover Man/ Blues GUIDE TO COLOUR CODING Etude / Some Day My Prince Will Come / Cubano Chant / Jordu / My Funny Valentine· Autumn leaves · You Look Good To Me / The Preacher BONUS TRACK : My Funny Valentine Audio CD’s only 2 disc - CD & DVD AFFINITY - Origins: The Baskervilles 1965 - SJPCD238 Christmas Ball, December 1965: She’s Not There/ I Feel Fine / It’s Good News Week/ Mr Tambourine Man / I Saw Her Standing There / Hallelujah I Love Her So / Freight Train/ Love Potion No9 / You like Me DVD only Too Much / Day Tripper / We Can Work It Out / Peggy Sue Got Married / I Can’t Get No Satisfaction/ Yesterday / Bumble Bee / Perfidia / Another Girl/ Summertime Blues / We Wish You A Merry Christ - mas/ Get Off My Cloud / Sweets For My Sweet/ DIGITAL only Hang On Sloopy / Get Off My Cloud· BONUS TRACKS: /Rehearsal, January 65: Mr Tambourine Man / Always Something There To Remind Me / You Like Me Too Much I Re - hearsal, June 65: Perfidia / Trains And Boats And Planes/ The last TIme /Chem - ADAM FAITH - I Survive - SJPDCD314 istry Society Party, December 65: Hey You’ve Got To Hide Your love Away/ -

Sdn Bhd (472852-W) Lot 2.-42, 2Nd Floor Low Yatt Plaza, No
PC Zone Computer Trading (M) Sdn Bhd (472852-W) Lot 2.-42, 2nd Floor Low Yatt Plaza, No. 7, Jalan 1/77 Off Jalan Bukit Bintang 55100 KL Tel:27156881, Fax:27156882 Email:[email protected] (Op.Hour 11.00am - 9.00pm) * Prices are subject to change without priority. Prices shown above are for Cash & Carry Only. Information provided is for reference only, actual procudt may be differ from batch to batch. PC Zone is not responsible for any typing error which has mistakenly made. Goods sold are non refundable or exchangeble.* Price list May be able to download from: http:/www.pczonecomputer.com or /my.hardwarezone.com/priceguide/ Intel Celeron Ghz Cache FSB RM Adata - Desktop RAM (Value) RM Kingston Desktop Ram - DDR2 (Value Series) RM Celeron 420 1.60 512K 800 125 Adata PC5400 DDR2 667 1GB 75 Kingston PC4200B DDR2 533 512MB 50 Celeron 430 1.80 512K 800 160 Adata PC5400 DDR2 667 2GB 145 Kingston PC4200B DDR2 533 1GB 71 Celeron 440 2.00 512K 800 185 Adata PC6400 DDR2 800 1GB 85 Kingston PC4200B DDR2 533 2 GB 150 Intel Celeron Dual Core Ghz Cache FSB RM Adata PC6400 DDR2 800 2GB 150 Kingston PC5400B DDR2 667 512MB 45 Celeron E1200 (Dual Core) 1.60 512K 800 195 Adata - Desktop RAM (Heatsink) RM Kingston PC5400B DDR2 667 1GB 65 Celeron E1400 (Dual Core) 2.00 512K 800 New Adata PC6400 DDR2 800 1GB 99 Kingston PC6400B DDR2 667 2 GB 150 Intel Pentium D Ghz Cache FSB RM Adata PC6400 DDR2 800 2x1GB 199 Kingston PC6400B DDR2 800 512MB 55 PentiumD 925 3.00 2x2MB 800 350 Adata PC6400 DDR2 800 2GB 349 Kingston PC6400B DDR2 800 1GB 80 PentiumD 945 -

NS4600 SR3 Compatibility List V1 1 -20111018
Promise ®®® SmartStor NS4600 SR3 Compatibility List V1.1 Promise ® SmartStor NS4600 SR3 Compatibility List V1.1 Date: 10/19/2011 Document History: V1.0: 1. First release ( Based on the original verification, SR1 , SR2 FW had a little bit minor changes) 2. Based on the original Compatibility List, added some new devices as below V1.1: 1. First release ( Based on the original verification, SR1 , SR2 FW had a little bit minor changes) 2. Based on the original Compatibility List, added some new devices as below (Highlighted them using blue-color in SR3 Compatibility List V1.1) Compatibility list included versions in this build are: Firmware: 02.01.0000.18 DEVICE COMPATIBILITY NOTE: Basic compatibility between the 3rd party device and Promise device has been verified in typical configurations, unless otherwise stated. The EOL of 3rd party device may not be in the current list, user can refer older version of Promise compatibility list to get those test result. DISCLAIMERS: Only the versions available at time of testing have been verified, as listed. Corner cases may exist depending on user configuration etc. Individual 3rd party units may exhibit problems that did not show up in through the Promise test process. Published performance is not assured with configuration deviations. Some listed devices may go EOL. AVL listing does not imply market availability. 1 of 15 Promise ®®® SmartStor NS4600 SR3 Compatibility List V1.1 1. Host systems (Ethernet ASIC on board) North Bridges/ NO. Vendor Model MB BIOS Ethernet ASIC South Bridges nVidia nForce -

Digital Archaeology: Rescuing Neglected and Damaged Data Resources I
Digital Archaeology: Rescuing Neglected and Damaged Data Resources I studies Digital Archaeology: Rescuing Neglected and Damaged Data Resources A JISC/NPO Study within the Electronic Libraries (eLib) Programme on the Preservation of Electronic Materials February 1999 Seamus Ross and Ann Gow Humanities Advanced Technology and Information Institute (HATII) University of Glasgow http://www.hatii.arts.gla.ac.uk/ eLib Study • P2 II Digital Archaeology: Rescuing Neglected and Damaged Data Resources © JISC 1999 ISBN 1 900508 51 6 Digital Archaeology: Rescuing Neglected and Damaged Data Resources was prepared as part of a programme of studies resulting from a workshop on the Long Term Preservation of Electronic Materials held at Warwick in November 1995. The programme of studies is guided by the Digital Archiving Working Group, which reports to the Management Committee of the National Preservation Office.The programme is administered by the British Library Research and Innovation Centre and funded by JISC, as part of the Electronic Libraries Programme. Whilst every effort has been made to ensure the accuracy of the contents of this publication, the publishers, the Electronic Libraries Programme, the Digital Archiving Working Group, the British Library Research and Innovation Centre and the Humanities Advanced Technology and Information Institute (HATII), University of Glasgow do not assume, and hereby disclaim, any liability to any party for loss or damage caused through errors or omissions in this publication, whether these errors or omissions result from accident, negligence or any other cause. The opinions expressed in this document are those of the authors and not necessarily those of the the publishers, the Electronic Libraries Programme, the Digital Archiving Working Group, the British Library Research and Innovation Centre, or the Humanities Advanced Technology and Information Institute (HATII), University of Glasgow.