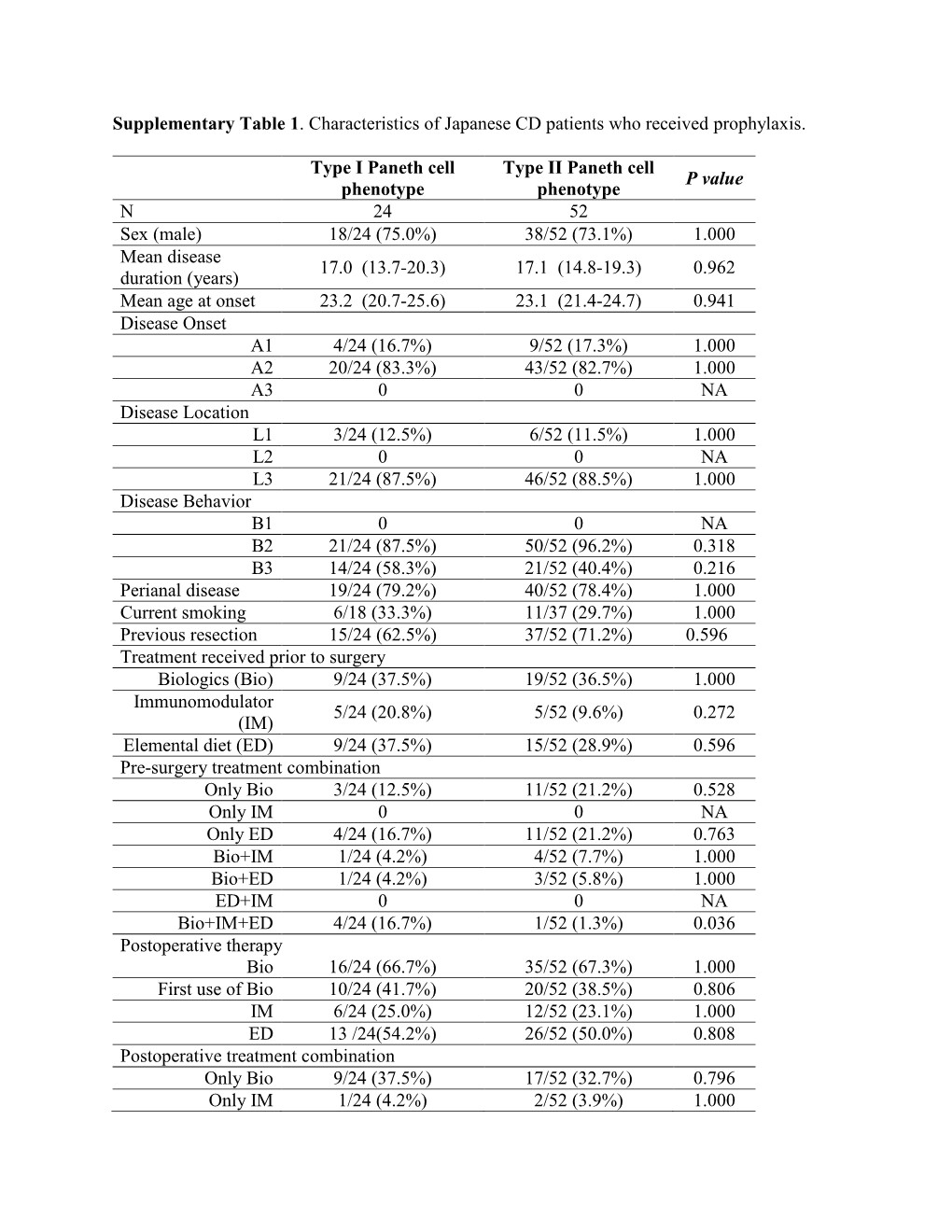
Supplementary Table 1. Characteristics of Japanese CD Patients Who Received Prophylaxis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Variation of SORBS1 Gene Is Associated with Glucose Homeostasis and Age at Onset of Diabetes: a Sapphire Cohort Study
www.nature.com/scientificreports OPEN Genetic variation of SORBS1 gene is associated with glucose homeostasis and age at onset of Received: 29 December 2017 Accepted: 19 June 2018 diabetes: A SAPPHIRe Cohort Study Published: xx xx xxxx Tien-Jyun Chang 1, Wen-Chang Wang2,3, Chao A. Hsiung3, Chih-Tsueng He4, Ming-Wei Lin5,6, Wayne Huey-Herng Sheu7,8,9, Yi-Cheng Chang1,10,11, Tom Quertermous12, Yii-Der Ida Chen13, Jerome I. Rotter13,14, Lee-Ming Chuang1,15 & The SAPPHIRe Study Group* The SORBS1 gene plays an important role in insulin signaling. We aimed to examine whether common single-nucleotide polymorphisms (SNPs) of SORBS1 are associated with prevalence and incidence of diabetes, age at onset of diabetes, and the related traits of glucose homeostasis. A total of 1135 siblings from 492 ethnic Chinese families were recruited at baseline, and 630 were followed up for 5.19 ± 0.96 years. Nine SNPs including rs7081076, rs2281939, rs3818540, rs2274490, rs61739184, rs726176, rs2296966, rs17849148, and rs3193970 were genotyped and examined. To deal with correlated data of subjects within the same families, the generalized estimating equations approach was applied throughout all association analyses. The GG genotype of rs2281939 was associated with a higher risk of diabetes at baseline, an earlier onset of diabetes, and higher steady-state plasma glucose levels in the modifed insulin suppression test. The minor allele T of rs2296966 was associated with higher prevalence and incidence of diabetes, an earlier onset of diabetes, and higher 2-h glucose during oral glucose tolerance test. These two SNPs revealed independent associations with age of diabetes onset as well as risk of diabetes at baseline. -

Vinexin Family (SORBS) Proteins Play Different Roles in Stiffness- Sensing and Contractile Force Generation Takafumi Ichikawa1,2, Masahiro Kita1, Tsubasa S
© 2017. Published by The Company of Biologists Ltd | Journal of Cell Science (2017) 130, 3517-3531 doi:10.1242/jcs.200691 RESEARCH ARTICLE Vinexin family (SORBS) proteins play different roles in stiffness- sensing and contractile force generation Takafumi Ichikawa1,2, Masahiro Kita1, Tsubasa S. Matsui3,4, Ayaka Ichikawa Nagasato1, Tomohiko Araki3, Shian-Huey Chiang5, Takuhito Sezaki1, Yasuhisa Kimura1, Kazumitsu Ueda1,2, Shinji Deguchi3,4, Alan R. Saltiel5,* and Noriyuki Kioka1,2,‡ ABSTRACT generating actin stress fibers (SFs) (Geiger et al., 2001). This Vinexin, c-Cbl associated protein (CAP) and Arg-binding protein 2 ‘ ’ (ArgBP2) constitute an adaptor protein family called the vinexin mechanical linkage acts as a molecular clutch to transmit the force (SORBS) family that is targeted to focal adhesions (FAs). Although derived from non-muscle myosin-II-dependent contraction to the numerous studies have focused on each of the SORBS proteins and ECM. Cells on more rigid substrates exert greater contractile forces partially elucidated their involvement in mechanotransduction, a than those on soft substrates (Hoffman et al., 2011; Roca-Cusachs comparative analysis of their function has not been well addressed. et al., 2012; LaCroix et al., 2015). These alterations can lead to Here, we established mouse embryonic fibroblasts that individually stiffness-dependent biochemical signals. Among the numerous FA scaffolding proteins, vinculin is one of expressed SORBS proteins and analysed their functions in an ‘ ’ identical cell context. Both vinexin-α and CAP co-localized with the main clutch molecules that can regulate force transmission. vinculin at FAs and promoted the appearance of vinculin-rich FAs, Vinculin consists of an N-terminal head region and a C-terminal tail α region separated by a flexible proline-rich linker region (Bakolitsa whereas ArgBP2 co-localized with -actinin at the proximal end of – FAs and punctate structures on actin stress fibers (SFs), and induced et al., 2004; Borgon et al., 2004). -

A Single Nucleotide Polymorphism in Exon 7 of Sorbin and SH3-Domain-Containing-1 (SORBS1) in Korean PCOS Patients
93-97 11/12/07 16:18 Page 93 MOLECULAR MEDICINE REPORTS 1: 93-97, 2008 93 A single nucleotide polymorphism in exon 7 of sorbin and SH3-domain-containing-1 (SORBS1) in Korean PCOS patients JUNG-MI PARK1, BON-HEE GU1, EUNG-JI LEE1, JAE-YOUNG KIM1, SUNG-WOO CHOI2 and KWANG-HYUN BAEK1 1Graduate School of Life Science and Biotechnology, Cell and Gene Therapy Research Institute, Pochon CHA University, CHA General Hospital, Seoul 135-081, Korea; 2Phillips Exeter Academy, Exeter, NH, USA Received October 19, 2007; Accepted November 20, 2007 Abstract. Patients with polycystic ovarian syndrome (PCOS) (3). It has been reported that approximately 4.9% of female are characterized by high levels of androgens, irregular or college students in Korea have exhibited PCOS (5). In women no menstrual cycle and increased hair growth. In addition, of other ethnic groups, that number has been reported as insulin resistance and glucose tolerance are caused in patients 4-12% (1,6,7). with PCOS. It was recently reported that PCOS in women is Recently, various single nucleotide polymorphisms (SNPs) associated with a single nucleotide polymorphism (SNP) of have been investigated by means of linkage and association genes, including the sorbin and SH3-domain-containing-1 studies in women with PCOS. A number of SNPs associated (SORBS1) gene involved in insulin resistance and glucose with PCOS have been reported (8-20), as have cases of PCOS uptake. SORBS1, which belongs to the sorbin homology related to the target genes of glucose uptake and insulin resis- (SoHo) family of proteins, becomes protein-protein interaction tance. -

Transcriptome Analysis of Adipose Tissues from Two Fat-Tailed Sheep
Li et al. BMC Genomics (2018) 19:338 https://doi.org/10.1186/s12864-018-4747-1 RESEARCHARTICLE Open Access Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition Baojun Li1, Liying Qiao1, Lixia An2, Weiwei Wang1, Jianhua Liu1, Youshe Ren1, Yangyang Pan1, Jiongjie Jing1 and Wenzhong Liu1* Abstract Background: The level of fat deposition in carcass is a crucial factor influencing meat quality. Guangling Large-Tailed (GLT) and Small-Tailed Han (STH) sheep are important local Chinese fat-tailed breeds that show distinct patterns of fat depots. To gain a better understanding of fat deposition, transcriptome profiles were determined by RNA-sequencing of perirenal, subcutaneous, and tail fat tissues from both the sheep breeds. The common highly expressed genes (co-genes) in all the six tissues, and the genes that were differentially expressed (DE genes) between these two breeds in the corresponding tissues were analyzed. Results: Approximately 47 million clean reads were obtained for each sample, and a total of 17,267 genes were annotated. Of the 47 highly expressed co-genes, FABP4, ADIPOQ, FABP5,andCD36 were the four most highly transcribed genes among all the known genes related to adipose deposition. FHC, FHC-pseudogene,and ZC3H10 were also highly expressed genes and could, thus, have roles in fat deposition. A total of 2091, 4233, and 4131 DE genes were identified in the perirenal, subcutaneous, and tail fat tissues between the GLT and STH breeds, respectively. Gene Ontology (GO) analysis showed that some DE genes were associated with adipose metabolism. -

Views for Entrez
BASIC RESEARCH www.jasn.org Phosphoproteomic Analysis Reveals Regulatory Mechanisms at the Kidney Filtration Barrier †‡ †| Markus M. Rinschen,* Xiongwu Wu,§ Tim König, Trairak Pisitkun,¶ Henning Hagmann,* † † † Caroline Pahmeyer,* Tobias Lamkemeyer, Priyanka Kohli,* Nicole Schnell, †‡ †† ‡‡ Bernhard Schermer,* Stuart Dryer,** Bernard R. Brooks,§ Pedro Beltrao, †‡ Marcus Krueger,§§ Paul T. Brinkkoetter,* and Thomas Benzing* *Department of Internal Medicine II, Center for Molecular Medicine, †Cologne Excellence Cluster on Cellular Stress | Responses in Aging-Associated Diseases, ‡Systems Biology of Ageing Cologne, Institute for Genetics, University of Cologne, Cologne, Germany; §Laboratory of Computational Biology, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland; ¶Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand; **Department of Biology and Biochemistry, University of Houston, Houston, Texas; ††Division of Nephrology, Baylor College of Medicine, Houston, Texas; ‡‡European Molecular Biology Laboratory–European Bioinformatics Institute, Hinxton, Cambridge, United Kingdom; and §§Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany ABSTRACT Diseases of the kidney filtration barrier are a leading cause of ESRD. Most disorders affect the podocytes, polarized cells with a limited capacity for self-renewal that require tightly controlled signaling to maintain their integrity, viability, and function. Here, we provide an atlas of in vivo phosphorylated, glomerulus- expressed -

Identification of Genetic Variations in CBL, SORBS1, CRK, and RHOQ
Genomics & Informatics Vol. 7(2) 53-56, June 2009 Identification of Genetic Variations in CBL, SORBS1, CRK, and RHOQ, Key Modulators in the CAP/TC10 Pathway of Insulin Signal Transduction, and Their Association with Type 2 Diabetes Mellitus in the Korean Population Kyung-Won Hong1, Hyun-Seok Jin1, Ji-Eun 1 2 2 Lim , Min Jin Go , Jong-Young Lee , Sue-Yun Introduction Hwang3, Hun Kuk Park1 and Bermseok Oh1* During recent years, major advances have been made in 1Department of Biomedical Engineering, School of Me- our understanding of insulin actions. In the cell, the rec- dicine, Kyung Hee University, Seoul 130-701, Korea, ognition of insulin by insulin receptor instigates phos- 2Center for Genome Science, National Institute of phorylation of several intracellular substrates, such as Health, Seoul 122-701, Korea, 3Graduate School of the insulin receptor substrates (IRS1-4) (White and Informatics and Biotechnology, Hankyung National Yenush, 1998), calcineurin B-like protein 1 (Cbl1) (Ribon University, Ansung 456-749, Korea and Saltiel, 1997), Src homology 2 domain-containing transforming protein (Shc) (Sasaoka et al., 1994), SH2B DOK Abstract adaptor protein 2 (SH2B2) (Liu et al., 2002), and p60 (Noguchi et al., 1999). Defects in genes involved in the Recent evidence has strongly suggested that the insulin signaling pathway are linked to the pathogenesis CAP/TC10 pathway is involved in the trafficking, dock- of type II diabetes mellitus (T2DM) (Zierath and Wall- ing, and fusion of vesicles containing the in- berg-Henriksson, 2002). sulin-responsive glucose transporter Glut4 to the plasma In the CAP/TC10 pathway, the insulin receptor re- membrane. -

Table of Content
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is © The Royal Society of Chemistry 2016 Supplemental Figures: Figure S1: Characterization of 96 Well Plates. (A) Image of 96 well glass-bottom plate used for cell culture and high content imaging. (B) Plot of atomic force microscope- determined stiffness of 5% and 10% acrylamide hydrogels, i.e. myogenic- and osteogenic-inducing hydrogels, respectively. (C) Schematic illustrating the diffusion based technique used to determine hydrogel thickness. Red circles indicate Texas Red- conjugated beads. (D) Plot of hydrogel thickness for 5% and 10% acrylamide hydrogels. Figure S2: CellProfiler Pipeline. (A) Nuclei are identified first from the DAPI channel. (B) Using the nuclei as seed regions, cell outlines are identified for each nucleus. (C) Representative images and their average nuclear and cytoplasmic fluorescence are shown to indicate marker expression and distribution information obtained through CellProlifer. Cells with high nuclear expression of the transcription factor are considered to have expressed and localized the factor correctly. Figure S3: ScanSite Analysis of Candidate Mechanosensors. (A) Plots of surface accessibility reveal regions corresponding to predicted MAPK1 domains. Green highlighted regions denote surface inaccessible binding sites predicted to bind MAPK1 domains. (B) List of sites and surface accessibility values for given predicted MAPK1 binding domains. Note that a surface accessible site has a value above 1 and a completely inaccessible site has a value of 0 in this analysis. Figure S4: Secondary Metrics from High Content Image Analysis. (A) Average cell area and (B) cell eccentricity, calculated as the ratio between the distance between the two foci of a fitted ellipse and the major axis length of the cell, are plotted as a function of siRNA treatment. -

PKB Is Required for Adipose Differentiation of Mouse Embryonic
Research Article 889 PKB␣ is required for adipose differentiation of mouse embryonic fibroblasts Anne Baudry, Zhong-Zhou Yang and Brian A. Hemmings* Friedrich Miescher Institute for Biomedical Research, Maulbeerstr. 66, CH-4058, Basel, Switzerland *Author for correspondence (e-mail: [email protected]) Accepted 14 November 2005 Journal of Cell Science 119, 889-897 Published by The Company of Biologists 2006 doi:10.1242/jcs.02792 Summary Protein kinase B␣ (PKB␣) is a key regulator of metabolism, downregulated in PKB␣-deficient MEFs but could be proliferation and differentiation. We have explored the role restored by expressing an active PKB␣ in the deficient cells. of PKB␣ in adipogenesis using wild-type and PKB␣- The level of lipocalin 2, renin 1 and receptor-activity- knockout mouse embryonic fibroblasts (MEFs) and show modifying protein 3 genes expressed by adipose cells was that lack of PKB␣ prevents MEF differentiation into also decreased in PKB␣-deficient MEFs, and are inhibited adipocytes. Expression of ectopic PKB␣ in PKB␣-deficient by LY294002 treatment during early adipocyte cells restores adipogenesis. We identified 80 genes whose differentiation of 3T3-L1 cells. The results underscore an expression was upregulated in wild-type MEFs during essential role for PKB␣ in the transcriptional program adipogenesis but whose expression was significantly required for adipogenesis. reduced in PKB␣-deficient MEFs under the same conditions. Significantly, the regulator of adipogenesis Key words: PKB␣, Adipocyte differentiation, Mouse embryonic Krüppel-like transcription factor 15 gene expression was fibroblasts, Microarray analysis Introduction many cellular processes stimulated by insulin and growth Adipogenesis is a complex process with multiple steps factors (Brazil and Hemmings, 2001; Lawlor and Alessi, 2001; including growth arrest, clonal expansion, withdrawal from the Whiteman et al., 2002). -

AFDN (Afadin, Adherens Junction Formation Factor)
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL INIST-CNRS Gene Section Review AFDN (afadin, adherens junction formation factor) Jean Loup Huret, Philippe Dessen [email protected] (JLH); UMR 1170 INSERM, Gustave Roussy, 114 rue Edouard Vaillant, 94805 Villejuif 94805, France (PHD) Published in Atlas Database: December 2018 Online updated version : http://AtlasGeneticsOncology.org/Genes/AF6ID6.html Printable original version : http://documents.irevues.inist.fr/bitstream/handle/2042/70479/12-2018-AF6ID6.pdf DOI: 10.4267/2042/70479 This article is an update of : Huret JL. MLLT4 (myeloid/lymphoid or mixed-lineage leukemia (trithorax). Atlas Genet Cytogenet Oncol Haematol 1997;1(2) This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2019 Atlas of Genetics and Cytogenetics in Oncology and Haematology Abstract Afadin, the protein coded by AFDN (6q27), also known as AF6 or MLLT4, is a cytoskeletal and junction-associated protein that links nectins, transmembrane proteins, to the F-actin (actin cytoskeleton) in a type of cell-cell junctions: the adherens junctions (AJs). Afadin plays an important role in AJs integrity and apical-basal polarity. There is growing evidence of it's role in carcinogenesis. Keywords Afadin; AFDN; AF6; MLLT4; Cytoskeleton; Cell- cell junctions; Adherens junctions; Apical-basal polarity; Epithelial-mesenchymal transition; Tight junctions; Mitotic spindle orientation; Migration; Nectin; Actin; Acute myeloid leukemia; Breast Figure 1 Localization of AFDN (also called AF6 or MLLT4) cancer; Colon cancer; Pancreatic cancer; - Courtesy Mariano Rocchi. Endometrial cancer; Gastric cancer; Osteosarcoma; Neurone synapse. DNA/RNA Identity Description HGNC (Hugo): AFDN The AFDN gene, located on 6q27, has a genomic size of 145,030 bases and encodes for several Location: 6q27 transcripts on the position chr6:167826991- Other names: MLLT4, AF6 167972020 in the + strand. -

Type 2 Diabetes Mellitus-Associated Transcriptome Alterations in Cortical
Bury et al. acta neuropathol commun (2021) 9:5 https://doi.org/10.1186/s40478-020-01109-y RESEARCH Open Access Type 2 diabetes mellitus-associated transcriptome alterations in cortical neurones and associated neurovascular unit cells in the ageing brain Joanna J. Bury1†, Annabelle Chambers1†, Paul R. Heath1, Paul G. Ince1, Pamela J. Shaw1, Fiona E. Matthews2, Carol Brayne3, Julie E. Simpson1† and Stephen B. Wharton1*† on behalf of the Cognitive Function and Ageing Study Abstract Type 2 diabetes mellitus (T2D), characterised by peripheral insulin resistance, is a risk factor for dementia. In addition to its contribution to small and large vessel disease, T2D may directly damage cells of the brain neurovascular unit. In this study, we investigated the transcriptomic changes in cortical neurones, and associated astrocytes and endothelial cells of the neurovascular unit, in the ageing brain. Neurone, astrocyte, and endothelial cell-enriched mRNA, obtained by immuno-laser capture microdissection of temporal cortex (Brodmann area 21/22) from 6 cases with self-reported T2D in the Cognitive Function and Ageing Study neuropathology cohort, and an equal number of age and sex- matched controls, was assessed by microarray analysis. Integrated Molecular Pathway Level Analysis was performed using the Kyoto Encyclopaedia of Genes and Genomes database on signifcantly diferentially expressed genes, defned as P < 0.05 and fold-change 1.2. Hub genes identifed from Weighted Gene Co-expression Network Analy- sis were validated in neurones using± the NanoString nCounter platform. The expression and cellular localisation of proteins encoded by selected candidate genes were confrmed by immunohistochemistry. 912, 2202, and 1227 genes were signifcantly diferentially expressed between cases with self-reported T2D and controls in neurones, astrocytes, and endothelial cells respectively. -

A Biochemical Landscape of A-To-I RNA Editing in the Human Brain Transcriptome
Downloaded from genome.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Resource A biochemical landscape of A-to-I RNA editing in the human brain transcriptome Masayuki Sakurai,1,4 Hiroki Ueda,1,4 Takanori Yano,1 Shunpei Okada,1 Hideki Terajima,1 Toutai Mitsuyama,2 Atsushi Toyoda,3 Asao Fujiyama,3 Hitomi Kawabata,1 and Tsutomu Suzuki1,5 1Department of Chemistry and Biotechnology, Graduate School of Engineering, University of Tokyo, Tokyo 113-8656, Japan; 2Computational Biology Research Center (CBRC), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo 135-0064, Japan; 3Comparative Genomics Laboratory, Center for Genetic Resource Information, National Institute of Genetics, Shizuoka 411-8540, Japan Inosine is an abundant RNA modification in the human transcriptome and is essential for many biological processes in modulating gene expression at the post-transcriptional level. Adenosine deaminases acting on RNA (ADARs) catalyze the hydrolytic deamination of adenosines to inosines (A-to-I editing) in double-stranded regions. We previously established a biochemical method called ‘‘inosine chemical erasing’’ (ICE) to directly identify inosines on RNA strands with high reliability. Here, we have applied the ICE method combined with deep sequencing (ICE-seq) to conduct an unbiased genome-wide screening of A-to-I editing sites in the transcriptome of human adult brain. Taken together with the sites identified by the conventional ICE method, we mapped 19,791 novel sites and newly found 1258 edited mRNAs, including 66 novel sites in coding regions, 41 of which cause altered amino acid assignment. ICE-seq detected novel editing sites in various repeat elements as well as in short hairpins. -
Genome-Wide Analysis of Glucocorticoid Receptor-Binding Sites in Myotubes Identifies Gene Networks Modulating Insulin Signaling
Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling Taiyi Kuoa,b, Michelle J. Lewb, Oleg Maybac, Charles A. Harrisd,e, Terence P. Speedc, and Jen-Chywan Wanga,b,1 aEndocrinology Graduate Program, bDepartment of Nutritional Science and Toxicology, and cDepartment of Statistics, University of California, Berkeley, CA 94720; dGladstone Institute for Cardiovascular Disease, San Francisco, CA 94158; and eDepartment of Medicine, University of California, San Francisco, CA 94143 Edited by Gordon L. Hager, National Cancer Institute, Bethesda, MD, and accepted by the Editorial Board May 30, 2012 (received for review July 15, 2011) Glucocorticoids elicit a variety of biological responses in skeletal and phosphorylate insulin receptor substrates (IRS) (14). Tyrosine- muscle, including inhibiting protein synthesis and insulin-stimu- phosphorylated IRS associate with insulin receptors (IR) and ac- lated glucose uptake and promoting proteolysis. Thus, excess or tivate signaling pathways (14). Mice treated with glucocorticoids chronic glucocorticoid exposure leads to muscle atrophy and insulin have reduced levels of tyrosine-phosphorylated IR and total IRS-1 resistance. Glucocorticoids propagate their signal mainly through in skeletal muscle (5). The activity of phosphatidylinositol 3-kinase glucocorticoid receptors (GR), which, upon binding to ligands, (PI3K) and Akt, two signaling molecules downstream of IR and translocate to the nucleus and bind to genomic glucocorticoid IRS-1, is also decreased (5). Glucocorticoids also reduce the activity response elements to regulate the transcription of nearby genes. of mammalian target of rapamycin (mTOR), a protein kinase Using a combination of chromatin immunoprecipitation sequenc- downstream of Akt and upstream of p70 S6 kinase (p70S6K) (15).