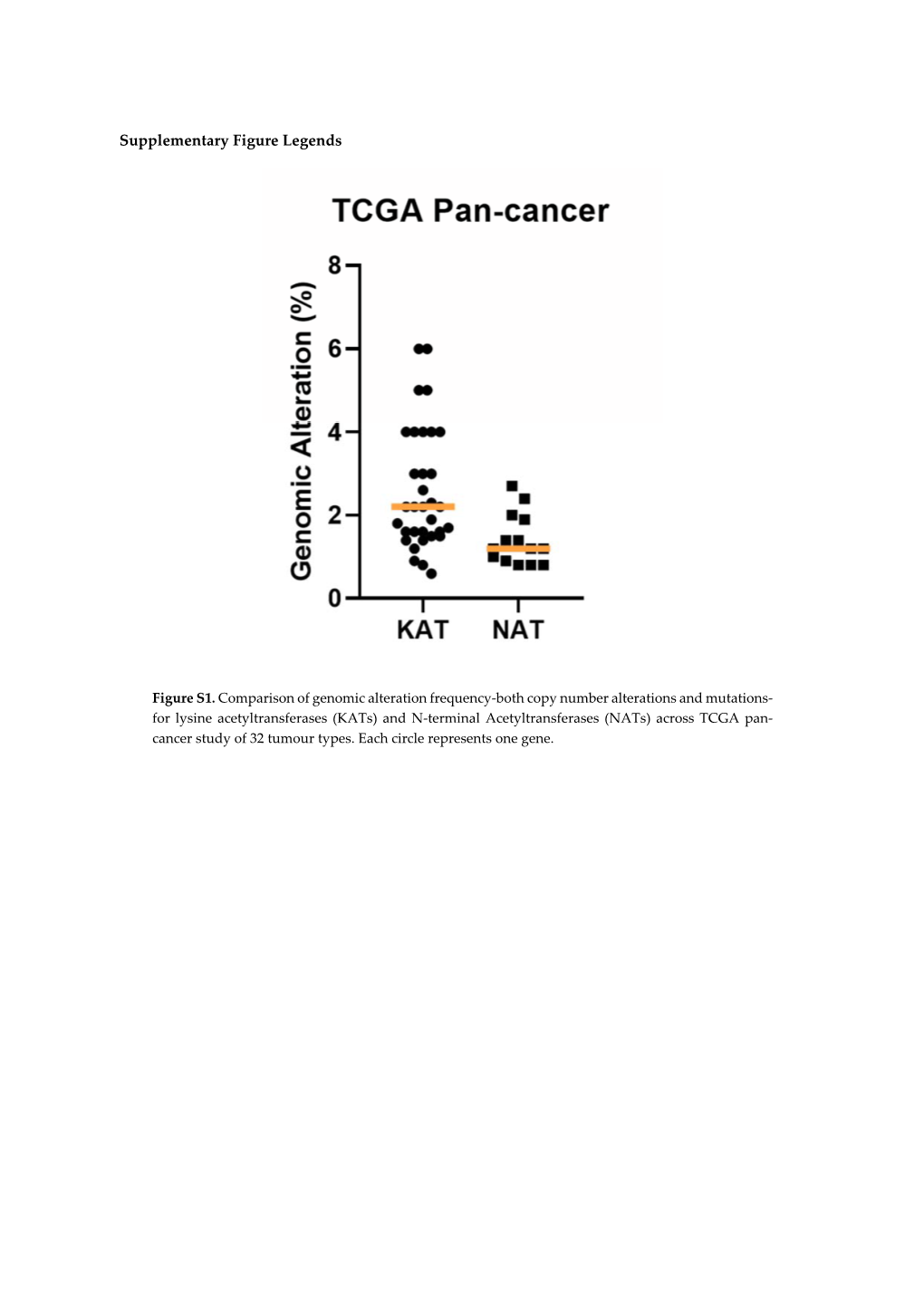
Supplementary Figure Legends
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Histone Isoform H2A1H Promotes Attainment of Distinct Physiological
Bhattacharya et al. Epigenetics & Chromatin (2017) 10:48 DOI 10.1186/s13072-017-0155-z Epigenetics & Chromatin RESEARCH Open Access Histone isoform H2A1H promotes attainment of distinct physiological states by altering chromatin dynamics Saikat Bhattacharya1,4,6, Divya Reddy1,4, Vinod Jani5†, Nikhil Gadewal3†, Sanket Shah1,4, Raja Reddy2,4, Kakoli Bose2,4, Uddhavesh Sonavane5, Rajendra Joshi5 and Sanjay Gupta1,4* Abstract Background: The distinct functional efects of the replication-dependent histone H2A isoforms have been dem- onstrated; however, the mechanistic basis of the non-redundancy remains unclear. Here, we have investigated the specifc functional contribution of the histone H2A isoform H2A1H, which difers from another isoform H2A2A3 in the identity of only three amino acids. Results: H2A1H exhibits varied expression levels in diferent normal tissues and human cancer cell lines (H2A1C in humans). It also promotes cell proliferation in a context-dependent manner when exogenously overexpressed. To uncover the molecular basis of the non-redundancy, equilibrium unfolding of recombinant H2A1H-H2B dimer was performed. We found that the M51L alteration at the H2A–H2B dimer interface decreases the temperature of melting of H2A1H-H2B by ~ 3 °C as compared to the H2A2A3-H2B dimer. This diference in the dimer stability is also refected in the chromatin dynamics as H2A1H-containing nucleosomes are more stable owing to M51L and K99R substitu- tions. Molecular dynamic simulations suggest that these substitutions increase the number of hydrogen bonds and hydrophobic interactions of H2A1H, enabling it to form more stable nucleosomes. Conclusion: We show that the M51L and K99R substitutions, besides altering the stability of histone–histone and histone–DNA complexes, have the most prominent efect on cell proliferation, suggesting that the nucleosome sta- bility is intimately linked with the physiological efects observed. -

Genome-Wide Analysis of 5-Hmc in the Peripheral Blood of Systemic Lupus Erythematosus Patients Using an Hmedip-Chip
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 35: 1467-1479, 2015 Genome-wide analysis of 5-hmC in the peripheral blood of systemic lupus erythematosus patients using an hMeDIP-chip WEIGUO SUI1*, QIUPEI TAN1*, MING YANG1, QIANG YAN1, HUA LIN1, MINGLIN OU1, WEN XUE1, JIEJING CHEN1, TONGXIANG ZOU1, HUANYUN JING1, LI GUO1, CUIHUI CAO1, YUFENG SUN1, ZHENZHEN CUI1 and YONG DAI2 1Guangxi Key Laboratory of Metabolic Diseases Research, Central Laboratory of Guilin 181st Hospital, Guilin, Guangxi 541002; 2Clinical Medical Research Center, the Second Clinical Medical College of Jinan University (Shenzhen People's Hospital), Shenzhen, Guangdong 518020, P.R. China Received July 9, 2014; Accepted February 27, 2015 DOI: 10.3892/ijmm.2015.2149 Abstract. Systemic lupus erythematosus (SLE) is a chronic, Introduction potentially fatal systemic autoimmune disease characterized by the production of autoantibodies against a wide range Systemic lupus erythematosus (SLE) is a typical systemic auto- of self-antigens. To investigate the role of the 5-hmC DNA immune disease, involving diffuse connective tissues (1) and modification with regard to the onset of SLE, we compared is characterized by immune inflammation. SLE has a complex the levels 5-hmC between SLE patients and normal controls. pathogenesis (2), involving genetic, immunologic and envi- Whole blood was obtained from patients, and genomic DNA ronmental factors. Thus, it may result in damage to multiple was extracted. Using the hMeDIP-chip analysis and valida- tissues and organs, especially the kidneys (3). SLE arises from tion by quantitative RT-PCR (RT-qPCR), we identified the a combination of heritable and environmental influences. differentially hydroxymethylated regions that are associated Epigenetics, the study of changes in gene expression with SLE. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Transcription-Factor Polymorphism Associated with Tissue-Specific Gene
bioRxiv preprint doi: https://doi.org/10.1101/405936; this version posted August 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. TF-TWAS: Transcription-factor polymorphism associated with tissue-specific gene expression Yi-Ching Tang 1, Assaf Gottlieb 1* 1 Center for precision health, School of Biomedical informatics, University of Texas Health Science Center at Houston, Houston, TX, 77030 * Corresponding author: Assaf Gottlieb, Center for Precision Health, School of Biomedical Informatics, University of Texas Health Science Center in Houston, 7000 Fannin st, Houston, TX, 77030 Email: [email protected] Tel: +1-(713) 5003698 Running Title: TF-TWAS: TF polymorphism affecting gene expression Keywords: “transcriptome-wide association studies”, “TWAS”, “Transcription Factor polymorphism” bioRxiv preprint doi: https://doi.org/10.1101/405936; this version posted August 31, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Transcriptional regulation is associated with a broad range of diseases. Methods associating genetic polymorphism with gene transcription levels offer key insights for understanding the transcriptional regulation plan. The majority of gene imputation methods focus on modeling polymorphism in the cis regions of the gene, partially owing to the large genetic search space. We hypothesize that polymorphism within transcription factors (TFs) may help explain transcription levels of their transcribed genes. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

HIST1H2AC Human Shrna Plasmid Kit (Locus ID 8334) Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TL312444 HIST1H2AC Human shRNA Plasmid Kit (Locus ID 8334) Product data: Product Type: shRNA Plasmids Product Name: HIST1H2AC Human shRNA Plasmid Kit (Locus ID 8334) Locus ID: 8334 Synonyms: dJ221C16.4; H2A/l; H2AFL; HIST1H2AC Vector: pGFP-C-shLenti (TR30023) Format: Lentiviral plasmids Components: HIST1H2AC - Human, 4 unique 29mer shRNA constructs in lentiviral GFP vector(Gene ID = 8334). 5µg purified plasmid DNA per construct Non-effective 29-mer scrambled shRNA cassette in pGFP-C-shLenti Vector, TR30021, included for free. RefSeq: NM_003512, NM_003512.1, NM_003512.2, NM_003512.3, BC085010, BC017379, BC050602, NM_003512.4 Summary: Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H2A family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6. [provided by RefSeq, Aug 2015] shRNA Design: These shRNA constructs were designed against multiple splice variants at this gene locus. -

Minichromosome Maintenance Proteins Are Direct Targets of the ATM and ATR Checkpoint Kinases
Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases David Cortez*†, Gloria Glick*, and Stephen J. Elledge‡§ *Department of Biochemistry, Vanderbilt University, Nashville, TN 37232; and ‡Department of Genetics, Center for Genetics and Genomics, and Howard Hughes Medical Institute, Harvard University Medical School, Boston, MA 02115 Contributed by Stephen J. Elledge, May 13, 2004 The minichromosome maintenance (MCM) 2-7 helicase complex unclear how they could be reassembled because replication functions to initiate and elongate replication forks. Cell cycle licensing is not allowed once S phase has begun (26). Continued checkpoint signaling pathways regulate DNA replication to main- MCM activity at a stalled replication fork may also be delete- tain genomic stability. We describe four lines of evidence that rious because it would expose extensive regions of single- ATM/ATR-dependent (ataxia-telangiectasia-mutated͞ATM- and stranded DNA, precisely the phenotype observed in S. cerevisiae Rad3-related) checkpoint pathways are directly linked to three checkpoint mutants treated with hydroxyurea (HU) (9). Fur- members of the MCM complex. First, ATM phosphorylates MCM3 thermore, short stretches of single-stranded DNA that might on S535 in response to ionizing radiation. Second, ATR phosphor- accumulate at stalled forks because of MCM action may be ylates MCM2 on S108 in response to multiple forms of DNA damage essential to recruiting the ATR-ATRIP complex and activating and stalling of replication forks. Third, ATR-interacting protein the S-phase checkpoint. In this study, we found four direct (ATRIP)-ATR interacts with MCM7. Fourth, reducing the amount of connections between the ATR͞ATM checkpoint pathways and MCM7 in cells disrupts checkpoint signaling and causes an intra- the MCM proteins. -

Cell Growth-Regulated Expression of Mammalian MCM5 and MCM6 Genes Mediated by the Transcription Factor E2F
Oncogene (1999) 18, 2299 ± 2309 ã 1999 Stockton Press All rights reserved 0950 ± 9232/99 $12.00 http://www.stockton-press.co.uk/onc Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F Kiyoshi Ohtani1, Ritsuko Iwanaga1, Masataka Nakamura*,1, Masa-aki Ikeda2, Norikazu Yabuta3, Hiromichi Tsuruga3 and Hiroshi Nojima3 1Human Gene Sciences Center, Tokyo Medical and Dental University, Tokyo 113-8510, Japan 2Department of Developmental Biology, Graduate School of Dentistry, Tokyo Medical and Dental University, Tokyo 113-8549, Japan; 3Department of Molecular Genetics, Research Institute for Microbial Diseases, Osaka University, Suita 565-0871, Japan Initiation of DNA replication requires the function of family (MCM2-7) that have been identi®ed in yeast, MCM gene products, which participate in ensuring that Xenopus, and human. Mcm proteins seem to regulate DNA replication occurs only once in the cell cycle. the initiation at the replication origin where the loading Expression of all mammalian genes of the MCM family of the proteins onto the origin recognition complex is induced by growth stimulation, unlike yeast, and the (ORC) is regulated by Cdc6 and cyclin-dependent mRNA levels peak at G1/S boundary. In this study, we kinases (Donovan et al., 1997; Tanaka et al., 1997). examined the transcriptional activities of isolated human However, the mechanism(s) by which Mcm proteins MCM gene promoters. Human MCM5 and MCM6 control the initiation of DNA replication remains promoters with mutation in the E2F sites failed in unclear. promoter regulation following serum stimulation and Xenopus Mcm proteins seem to be able to access exogenous E2F expression. -

Intrinsic Disorder in PTEN and Its Interactome Confers Structural Plasticity and 63
OPEN Intrinsic Disorder in PTEN and its SUBJECT AREAS: Interactome Confers Structural Plasticity CELLULAR SIGNALLING NETWORKS and Functional Versatility CANCER GENOMICS Prerna Malaney1*, Ravi Ramesh Pathak1*, Bin Xue3, Vladimir N. Uversky3,4,5 & Vrushank Dave´1,2 SYSTEMS ANALYSIS GENE REGULATORY NETWORKS 1Morsani College of Medicine, Department of Pathology and Cell Biology, 2Department of Molecular Oncology, H. Lee Moffitt Cancer Center and Research Institute, 3Department of Molecular Medicine, 4USF Health Byrd Alzheimer’s Research Institute, Received University of South Florida, Tampa, FL, 33612, USA, 5Institute for Biological Instrumentation, Russian Academy of Sciences, 142290 11 March 2013 Pushchino, Moscow Region, Russia. Accepted 3 June 2013 IDPs, while structurally poor, are functionally rich by virtue of their flexibility and modularity. However, how mutations in IDPs elicit diseases, remain elusive. Herein, we have identified tumor suppressor PTEN as Published an intrinsically disordered protein (IDP) and elucidated the molecular principles by which its intrinsically 20 June 2013 disordered region (IDR) at the carboxyl-terminus (C-tail) executes its functions. Post-translational modifications, conserved eukaryotic linear motifs and molecular recognition features present in the C-tail IDR enhance PTEN’s protein-protein interactions that are required for its myriad cellular functions. PTEN primary and secondary interactomes are also enriched in IDPs, most being cancer related, revealing that Correspondence and PTEN functions emanate from and are nucleated by the C-tail IDR, which form pliable network-hubs. requests for materials Together, PTEN higher order functional networks operate via multiple IDP-IDP interactions facilitated by should be addressed to its C-tail IDR. Targeting PTEN IDR and its interaction hubs emerges as a new paradigm for treatment of V.D. -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

Bioinformatics-Based Screening of Key Genes for Transformation of Liver
Jiang et al. J Transl Med (2020) 18:40 https://doi.org/10.1186/s12967-020-02229-8 Journal of Translational Medicine RESEARCH Open Access Bioinformatics-based screening of key genes for transformation of liver cirrhosis to hepatocellular carcinoma Chen Hao Jiang1,2, Xin Yuan1,2, Jiang Fen Li1,2, Yu Fang Xie1,2, An Zhi Zhang1,2, Xue Li Wang1,2, Lan Yang1,2, Chun Xia Liu1,2, Wei Hua Liang1,2, Li Juan Pang1,2, Hong Zou1,2, Xiao Bin Cui1,2, Xi Hua Shen1,2, Yan Qi1,2, Jin Fang Jiang1,2, Wen Yi Gu4, Feng Li1,2,3 and Jian Ming Hu1,2* Abstract Background: Hepatocellular carcinoma (HCC) is the most common type of liver tumour, and is closely related to liver cirrhosis. Previous studies have focussed on the pathogenesis of liver cirrhosis developing into HCC, but the molecular mechanism remains unclear. The aims of the present study were to identify key genes related to the transformation of cirrhosis into HCC, and explore the associated molecular mechanisms. Methods: GSE89377, GSE17548, GSE63898 and GSE54236 mRNA microarray datasets from Gene Expression Omni- bus (GEO) were analysed to obtain diferentially expressed genes (DEGs) between HCC and liver cirrhosis tissues, and network analysis of protein–protein interactions (PPIs) was carried out. String and Cytoscape were used to analyse modules and identify hub genes, Kaplan–Meier Plotter and Oncomine databases were used to explore relationships between hub genes and disease occurrence, development and prognosis of HCC, and the molecular mechanism of the main hub gene was probed using Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway analysis. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine