A Novel Encoding Algorithm for Textual Data Compression
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Data Compression: Dictionary-Based Coding 2 / 37 Dictionary-Based Coding Dictionary-Based Coding
Dictionary-based Coding already coded not yet coded search buffer look-ahead buffer cursor (N symbols) (L symbols) We know the past but cannot control it. We control the future but... Last Lecture Last Lecture: Predictive Lossless Coding Predictive Lossless Coding Simple and effective way to exploit dependencies between neighboring symbols / samples Optimal predictor: Conditional mean (requires storage of large tables) Affine and Linear Prediction Simple structure, low-complex implementation possible Optimal prediction parameters are given by solution of Yule-Walker equations Works very well for real signals (e.g., audio, images, ...) Efficient Lossless Coding for Real-World Signals Affine/linear prediction (often: block-adaptive choice of prediction parameters) Entropy coding of prediction errors (e.g., arithmetic coding) Using marginal pmf often already yields good results Can be improved by using conditional pmfs (with simple conditions) Heiko Schwarz (Freie Universität Berlin) — Data Compression: Dictionary-based Coding 2 / 37 Dictionary-based Coding Dictionary-Based Coding Coding of Text Files Very high amount of dependencies Affine prediction does not work (requires linear dependencies) Higher-order conditional coding should work well, but is way to complex (memory) Alternative: Do not code single characters, but words or phrases Example: English Texts Oxford English Dictionary lists less than 230 000 words (including obsolete words) On average, a word contains about 6 characters Average codeword length per character would be limited by 1 -

Package 'Brotli'
Package ‘brotli’ May 13, 2018 Type Package Title A Compression Format Optimized for the Web Version 1.2 Description A lossless compressed data format that uses a combination of the LZ77 algorithm and Huffman coding. Brotli is similar in speed to deflate (gzip) but offers more dense compression. License MIT + file LICENSE URL https://tools.ietf.org/html/rfc7932 (spec) https://github.com/google/brotli#readme (upstream) http://github.com/jeroen/brotli#read (devel) BugReports http://github.com/jeroen/brotli/issues VignetteBuilder knitr, R.rsp Suggests spelling, knitr, R.rsp, microbenchmark, rmarkdown, ggplot2 RoxygenNote 6.0.1 Language en-US NeedsCompilation yes Author Jeroen Ooms [aut, cre] (<https://orcid.org/0000-0002-4035-0289>), Google, Inc [aut, cph] (Brotli C++ library) Maintainer Jeroen Ooms <[email protected]> Repository CRAN Date/Publication 2018-05-13 20:31:43 UTC R topics documented: brotli . .2 Index 4 1 2 brotli brotli Brotli Compression Description Brotli is a compression algorithm optimized for the web, in particular small text documents. Usage brotli_compress(buf, quality = 11, window = 22) brotli_decompress(buf) Arguments buf raw vector with data to compress/decompress quality value between 0 and 11 window log of window size Details Brotli decompression is at least as fast as for gzip while significantly improving the compression ratio. The price we pay is that compression is much slower than gzip. Brotli is therefore most effective for serving static content such as fonts and html pages. For binary (non-text) data, the compression ratio of Brotli usually does not beat bz2 or xz (lzma), however decompression for these algorithms is too slow for browsers in e.g. -
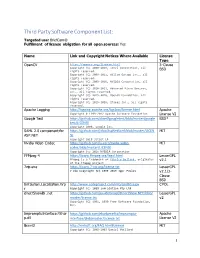
Third Party Software Component List: Targeted Use: Briefcam® Fulfillment of License Obligation for All Open Sources: Yes
Third Party Software Component List: Targeted use: BriefCam® Fulfillment of license obligation for all open sources: Yes Name Link and Copyright Notices Where Available License Type OpenCV https://opencv.org/license.html 3-Clause Copyright (C) 2000-2019, Intel Corporation, all BSD rights reserved. Copyright (C) 2009-2011, Willow Garage Inc., all rights reserved. Copyright (C) 2009-2016, NVIDIA Corporation, all rights reserved. Copyright (C) 2010-2013, Advanced Micro Devices, Inc., all rights reserved. Copyright (C) 2015-2016, OpenCV Foundation, all rights reserved. Copyright (C) 2015-2016, Itseez Inc., all rights reserved. Apache Logging http://logging.apache.org/log4cxx/license.html Apache Copyright © 1999-2012 Apache Software Foundation License V2 Google Test https://github.com/abseil/googletest/blob/master/google BSD* test/LICENSE Copyright 2008, Google Inc. SAML 2.0 component for https://github.com/jitbit/AspNetSaml/blob/master/LICEN MIT ASP.NET SE Copyright 2018 Jitbit LP Nvidia Video Codec https://github.com/lu-zero/nvidia-video- MIT codec/blob/master/LICENSE Copyright (c) 2016 NVIDIA Corporation FFMpeg 4 https://www.ffmpeg.org/legal.html LesserGPL FFmpeg is a trademark of Fabrice Bellard, originator v2.1 of the FFmpeg project 7zip.exe https://www.7-zip.org/license.txt LesserGPL 7-Zip Copyright (C) 1999-2019 Igor Pavlov v2.1/3- Clause BSD Infralution.Localization.Wp http://www.codeproject.com/info/cpol10.aspx CPOL f Copyright (C) 2018 Infralution Pty Ltd directShowlib .net https://github.com/pauldotknopf/DirectShow.NET/blob/ LesserGPL -

Video Codec Requirements and Evaluation Methodology
Video Codec Requirements 47pt 30pt and Evaluation Methodology Color::white : LT Medium Font to be used by customers and : Arial www.huawei.com draft-filippov-netvc-requirements-01 Alexey Filippov, Huawei Technologies 35pt Contents Font to be used by customers and partners : • An overview of applications • Requirements 18pt • Evaluation methodology Font to be used by customers • Conclusions and partners : Slide 2 Page 2 35pt Applications Font to be used by customers and partners : • Internet Protocol Television (IPTV) • Video conferencing 18pt • Video sharing Font to be used by customers • Screencasting and partners : • Game streaming • Video monitoring / surveillance Slide 3 35pt Internet Protocol Television (IPTV) Font to be used by customers and partners : • Basic requirements: . Random access to pictures 18pt Random Access Period (RAP) should be kept small enough (approximately, 1-15 seconds); Font to be used by customers . Temporal (frame-rate) scalability; and partners : . Error robustness • Optional requirements: . resolution and quality (SNR) scalability Slide 4 35pt Internet Protocol Television (IPTV) Font to be used by customers and partners : Resolution Frame-rate, fps Picture access mode 2160p (4K),3840x2160 60 RA 18pt 1080p, 1920x1080 24, 50, 60 RA 1080i, 1920x1080 30 (60 fields per second) RA Font to be used by customers and partners : 720p, 1280x720 50, 60 RA 576p (EDTV), 720x576 25, 50 RA 576i (SDTV), 720x576 25, 30 RA 480p (EDTV), 720x480 50, 60 RA 480i (SDTV), 720x480 25, 30 RA Slide 5 35pt Video conferencing Font to be used by customers and partners : • Basic requirements: . Delay should be kept as low as possible 18pt The preferable and maximum delay values should be less than 100 ms and 350 ms, respectively Font to be used by customers . -

Compressed Transitive Delta Encoding 1. Introduction
Compressed Transitive Delta Encoding Dana Shapira Department of Computer Science Ashkelon Academic College Ashkelon 78211, Israel [email protected] Abstract Given a source file S and two differencing files ∆(S; T ) and ∆(T;R), where ∆(X; Y ) is used to denote the delta file of the target file Y with respect to the source file X, the objective is to be able to construct R. This is intended for the scenario of upgrading soft- ware where intermediate releases are missing, or for the case of file system backups, where non consecutive versions must be recovered. The traditional way is to decompress ∆(S; T ) in order to construct T and then apply ∆(T;R) on T and obtain R. The Compressed Transitive Delta Encoding (CTDE) paradigm, introduced in this paper, is to construct a delta file ∆(S; R) working directly on the two given delta files, ∆(S; T ) and ∆(T;R), without any decompression or the use of the base file S. A new algorithm for solving CTDE is proposed and its compression performance is compared against the traditional \double delta decompression". Not only does it use constant additional space, as opposed to the traditional method which uses linear additional memory storage, but experiments show that the size of the delta files involved is reduced by 15% on average. 1. Introduction Differential file compression represents a target file T with respect to a source file S. That is, both the encoder and decoder have available identical copies of S. A new file T is encoded and subsequently decoded by making use of S. -

Arithmetic Coding
Arithmetic Coding Arithmetic coding is the most efficient method to code symbols according to the probability of their occurrence. The average code length corresponds exactly to the possible minimum given by information theory. Deviations which are caused by the bit-resolution of binary code trees do not exist. In contrast to a binary Huffman code tree the arithmetic coding offers a clearly better compression rate. Its implementation is more complex on the other hand. In arithmetic coding, a message is encoded as a real number in an interval from one to zero. Arithmetic coding typically has a better compression ratio than Huffman coding, as it produces a single symbol rather than several separate codewords. Arithmetic coding differs from other forms of entropy encoding such as Huffman coding in that rather than separating the input into component symbols and replacing each with a code, arithmetic coding encodes the entire message into a single number, a fraction n where (0.0 ≤ n < 1.0) Arithmetic coding is a lossless coding technique. There are a few disadvantages of arithmetic coding. One is that the whole codeword must be received to start decoding the symbols, and if there is a corrupt bit in the codeword, the entire message could become corrupt. Another is that there is a limit to the precision of the number which can be encoded, thus limiting the number of symbols to encode within a codeword. There also exist many patents upon arithmetic coding, so the use of some of the algorithms also call upon royalty fees. Arithmetic coding is part of the JPEG data format. -

The Basic Principles of Data Compression
The Basic Principles of Data Compression Author: Conrad Chung, 2BrightSparks Introduction Internet users who download or upload files from/to the web, or use email to send or receive attachments will most likely have encountered files in compressed format. In this topic we will cover how compression works, the advantages and disadvantages of compression, as well as types of compression. What is Compression? Compression is the process of encoding data more efficiently to achieve a reduction in file size. One type of compression available is referred to as lossless compression. This means the compressed file will be restored exactly to its original state with no loss of data during the decompression process. This is essential to data compression as the file would be corrupted and unusable should data be lost. Another compression category which will not be covered in this article is “lossy” compression often used in multimedia files for music and images and where data is discarded. Lossless compression algorithms use statistic modeling techniques to reduce repetitive information in a file. Some of the methods may include removal of spacing characters, representing a string of repeated characters with a single character or replacing recurring characters with smaller bit sequences. Advantages/Disadvantages of Compression Compression of files offer many advantages. When compressed, the quantity of bits used to store the information is reduced. Files that are smaller in size will result in shorter transmission times when they are transferred on the Internet. Compressed files also take up less storage space. File compression can zip up several small files into a single file for more convenient email transmission. -

(A/V Codecs) REDCODE RAW (.R3D) ARRIRAW
What is a Codec? Codec is a portmanteau of either "Compressor-Decompressor" or "Coder-Decoder," which describes a device or program capable of performing transformations on a data stream or signal. Codecs encode a stream or signal for transmission, storage or encryption and decode it for viewing or editing. Codecs are often used in videoconferencing and streaming media solutions. A video codec converts analog video signals from a video camera into digital signals for transmission. It then converts the digital signals back to analog for display. An audio codec converts analog audio signals from a microphone into digital signals for transmission. It then converts the digital signals back to analog for playing. The raw encoded form of audio and video data is often called essence, to distinguish it from the metadata information that together make up the information content of the stream and any "wrapper" data that is then added to aid access to or improve the robustness of the stream. Most codecs are lossy, in order to get a reasonably small file size. There are lossless codecs as well, but for most purposes the almost imperceptible increase in quality is not worth the considerable increase in data size. The main exception is if the data will undergo more processing in the future, in which case the repeated lossy encoding would damage the eventual quality too much. Many multimedia data streams need to contain both audio and video data, and often some form of metadata that permits synchronization of the audio and video. Each of these three streams may be handled by different programs, processes, or hardware; but for the multimedia data stream to be useful in stored or transmitted form, they must be encapsulated together in a container format. -

Encryption Introduction to Using 7-Zip
IT Services Training Guide Encryption Introduction to using 7-Zip It Services Training Team The University of Manchester email: [email protected] www.itservices.manchester.ac.uk/trainingcourses/coursesforstaff Version: 5.3 Training Guide Introduction to Using 7-Zip Page 2 IT Services Training Introduction to Using 7-Zip Table of Contents Contents Introduction ......................................................................................................................... 4 Compress/encrypt individual files ....................................................................................... 5 Email compressed/encrypted files ....................................................................................... 8 Decrypt an encrypted file ..................................................................................................... 9 Create a self-extracting encrypted file .............................................................................. 10 Decrypt/un-zip a file .......................................................................................................... 14 APPENDIX A Downloading and installing 7-Zip ................................................................. 15 Help and Further Reference ............................................................................................... 18 Page 3 Training Guide Introduction to Using 7-Zip Introduction 7-Zip is an application that allows you to: Compress a file – for example a file that is 5MB can be compressed to 3MB Secure the -

Implementing Compression on Distributed Time Series Database
Implementing compression on distributed time series database Michael Burman School of Science Thesis submitted for examination for the degree of Master of Science in Technology. Espoo 05.11.2017 Supervisor Prof. Kari Smolander Advisor Mgr. Jiri Kremser Aalto University, P.O. BOX 11000, 00076 AALTO www.aalto.fi Abstract of the master’s thesis Author Michael Burman Title Implementing compression on distributed time series database Degree programme Major Computer Science Code of major SCI3042 Supervisor Prof. Kari Smolander Advisor Mgr. Jiri Kremser Date 05.11.2017 Number of pages 70+4 Language English Abstract Rise of microservices and distributed applications in containerized deployments are putting increasing amount of burden to the monitoring systems. They push the storage requirements to provide suitable performance for large queries. In this paper we present the changes we made to our distributed time series database, Hawkular-Metrics, and how it stores data more effectively in the Cassandra. We show that using our methods provides significant space savings ranging from 50 to 95% reduction in storage usage, while reducing the query times by over 90% compared to the nominal approach when using Cassandra. We also provide our unique algorithm modified from Gorilla compression algorithm that we use in our solution, which provides almost three times the throughput in compression with equal compression ratio. Keywords timeseries compression performance storage Aalto-yliopisto, PL 11000, 00076 AALTO www.aalto.fi Diplomityön tiivistelmä Tekijä Michael Burman Työn nimi Pakkausmenetelmät hajautetussa aikasarjatietokannassa Koulutusohjelma Pääaine Computer Science Pääaineen koodi SCI3042 Työn valvoja ja ohjaaja Prof. Kari Smolander Päivämäärä 05.11.2017 Sivumäärä 70+4 Kieli Englanti Tiivistelmä Hajautettujen järjestelmien yleistyminen on aiheuttanut valvontajärjestelmissä tiedon määrän kasvua, sillä aikasarjojen määrä on kasvanut ja niihin talletetaan useammin tietoa. -

Metadefender Core V4.12.2
MetaDefender Core v4.12.2 © 2018 OPSWAT, Inc. All rights reserved. OPSWAT®, MetadefenderTM and the OPSWAT logo are trademarks of OPSWAT, Inc. All other trademarks, trade names, service marks, service names, and images mentioned and/or used herein belong to their respective owners. Table of Contents About This Guide 13 Key Features of Metadefender Core 14 1. Quick Start with Metadefender Core 15 1.1. Installation 15 Operating system invariant initial steps 15 Basic setup 16 1.1.1. Configuration wizard 16 1.2. License Activation 21 1.3. Scan Files with Metadefender Core 21 2. Installing or Upgrading Metadefender Core 22 2.1. Recommended System Requirements 22 System Requirements For Server 22 Browser Requirements for the Metadefender Core Management Console 24 2.2. Installing Metadefender 25 Installation 25 Installation notes 25 2.2.1. Installing Metadefender Core using command line 26 2.2.2. Installing Metadefender Core using the Install Wizard 27 2.3. Upgrading MetaDefender Core 27 Upgrading from MetaDefender Core 3.x 27 Upgrading from MetaDefender Core 4.x 28 2.4. Metadefender Core Licensing 28 2.4.1. Activating Metadefender Licenses 28 2.4.2. Checking Your Metadefender Core License 35 2.5. Performance and Load Estimation 36 What to know before reading the results: Some factors that affect performance 36 How test results are calculated 37 Test Reports 37 Performance Report - Multi-Scanning On Linux 37 Performance Report - Multi-Scanning On Windows 41 2.6. Special installation options 46 Use RAMDISK for the tempdirectory 46 3. Configuring Metadefender Core 50 3.1. Management Console 50 3.2. -

A Novel Coding Architecture for Multi-Line Lidar Point Clouds
This article has been accepted for inclusion in a future issue of this journal. Content is final as presented, with the exception of pagination. IEEE TRANSACTIONS ON INTELLIGENT TRANSPORTATION SYSTEMS 1 A Novel Coding Architecture for Multi-Line LiDAR Point Clouds Based on Clustering and Convolutional LSTM Network Xuebin Sun , Sukai Wang , Graduate Student Member, IEEE, and Ming Liu , Senior Member, IEEE Abstract— Light detection and ranging (LiDAR) plays an preservation of historical relics, 3D sensing for smart city, indispensable role in autonomous driving technologies, such as well as autonomous driving. Especially for autonomous as localization, map building, navigation and object avoidance. driving systems, LiDAR sensors play an indispensable role However, due to the vast amount of data, transmission and storage could become an important bottleneck. In this article, in a large number of key techniques, such as simultaneous we propose a novel compression architecture for multi-line localization and mapping (SLAM) [1], path planning [2], LiDAR point cloud sequences based on clustering and convolu- obstacle avoidance [3], and navigation. A point cloud consists tional long short-term memory (LSTM) networks. LiDAR point of a set of individual 3D points, in accordance with one or clouds are structured, which provides an opportunity to convert more attributes (color, reflectance, surface normal, etc). For the 3D data to 2D array, represented as range images. Thus, we cast the 3D point clouds compression as a range image instance, the Velodyne HDL-64E LiDAR sensor generates a sequence compression problem. Inspired by the high efficiency point cloud of up to 2.2 billion points per second, with a video coding (HEVC) algorithm, we design a novel compression range of up to 120 m.