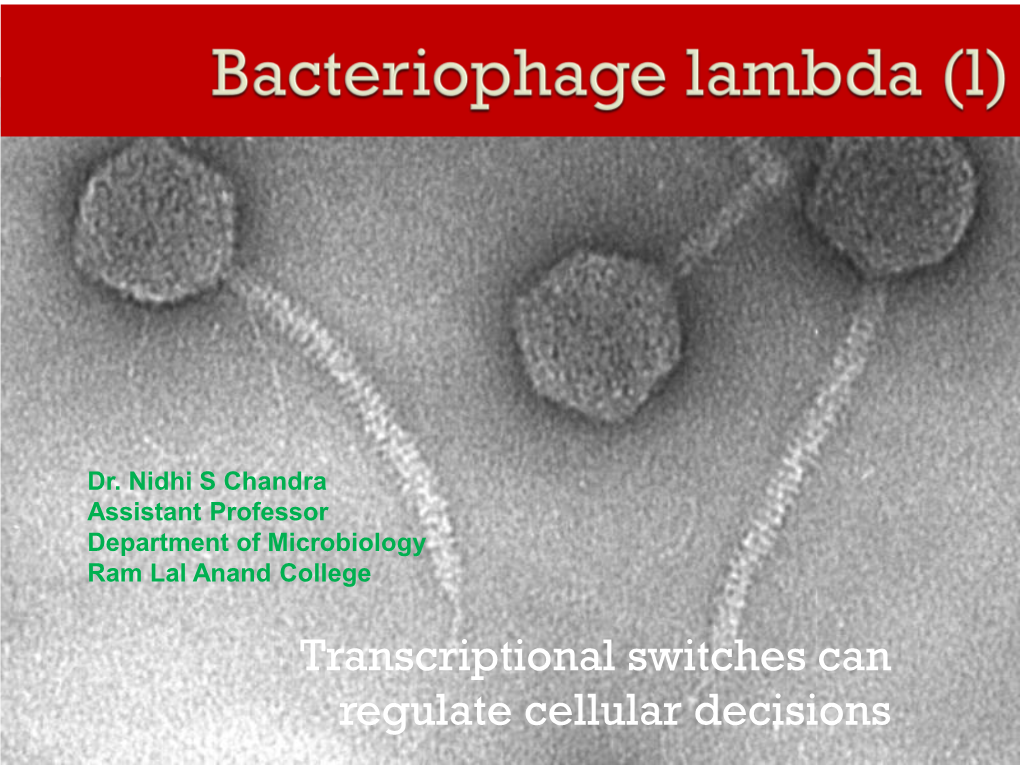
Bacteriophage Lambda
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

P1 Bacteriophage and Tol System Mutants
P1 BACTERIOPHAGE AND TOL SYSTEM MUTANTS Cari L. Smerk A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2007 Committee: Ray A. Larsen, Advisor Tami C. Steveson Paul A. Moore Lee A. Meserve ii ABSTRACT Dr. Ray A. Larsen, Advisor The integrity of the outer membrane of Gram negative bacteria is dependent upon proteins of the Tol system, which transduce cytoplasmic-membrane derived energy to as yet unidentified outer membrane targets (Vianney et al., 1996). Mutations affecting the Tol system of Escherichia coli render the cells resistant to a bacteriophage called P1 by blocking the phage maturation process in some way. This does not involve outer membrane interactions, as a mutant in the energy transucer (TolA) retained wild type levels of phage sensitivity. Conversely, mutations affecting the energy harvesting complex component, TolQ, were resistant to lysis by bacteriophage P1. Further characterization of specific Tol system mutants suggested that phage maturation was not coupled to energy transduction, nor to infection of the cells by the phage. Quantification of the number of phage produced by strains lacking this protein also suggests that the maturation of P1 phage requires conditions influenced by TolQ. This study aims to identify the role that the TolQ protein plays in the phage maturation process. Strains of cells were inoculated with bacteriophage P1 and the resulting production by the phage of viable progeny were determined using one step growth curves (Ellis and Delbruck, 1938). Strains that were lacking the TolQ protein rendered P1 unable to produce the characteristic burst of progeny phage after a single generation of phage. -

Protists and the Wild, Wild West of Gene Expression
MI70CH09-Keeling ARI 3 August 2016 18:22 ANNUAL REVIEWS Further Click here to view this article's online features: • Download figures as PPT slides • Navigate linked references • Download citations Protists and the Wild, Wild • Explore related articles • Search keywords West of Gene Expression: New Frontiers, Lawlessness, and Misfits David Roy Smith1 and Patrick J. Keeling2 1Department of Biology, University of Western Ontario, London, Ontario, Canada N6A 5B7; email: [email protected] 2Canadian Institute for Advanced Research, Department of Botany, University of British Columbia, Vancouver, British Columbia, Canada V6T 1Z4; email: [email protected] Annu. Rev. Microbiol. 2016. 70:161–78 Keywords First published online as a Review in Advance on constructive neutral evolution, mitochondrial transcription, plastid June 17, 2016 transcription, posttranscriptional processing, RNA editing, trans-splicing The Annual Review of Microbiology is online at micro.annualreviews.org Abstract This article’s doi: The DNA double helix has been called one of life’s most elegant structures, 10.1146/annurev-micro-102215-095448 largely because of its universality, simplicity, and symmetry. The expression Annu. Rev. Microbiol. 2016.70:161-178. Downloaded from www.annualreviews.org Copyright c 2016 by Annual Reviews. Access provided by University of British Columbia on 09/24/17. For personal use only. of information encoded within DNA, however, can be far from simple or All rights reserved symmetric and is sometimes surprisingly variable, convoluted, and wantonly inefficient. Although exceptions to the rules exist in certain model systems, the true extent to which life has stretched the limits of gene expression is made clear by nonmodel systems, particularly protists (microbial eukary- otes). -

|||||||III US005478731A United States Patent (19) 11 Patent Number: 5,478,731
|||||||III US005478731A United States Patent (19) 11 Patent Number: 5,478,731 Short 45 Date of Patent: Dec. 26,9 1995 54). POLYCOS VECTORS 56 References Cited 75) Inventor: Jay M. Short, Encinitas, Calif. PUBLICATIONS (73 Assignee: Stratagene, La Jolla, Calif. Ahmed (1989), Gene 75:315-321. 21 Appl. No.: 133,179 Evans et al. (1989), Gene 79:9-20. 22) PCT Filed: Apr. 10, 1992 Primary Examiner-Mindy B. Fleisher Assistant Examiner-Philip W. Carter 86 PCT No.: PCT/US92/03012 Attorney, Agent, or Firm-Albert P. Halluin; Pennie & S371 Date: Oct. 12, 1993 Edmonds S 102(e) Date: Oct. 12, 1993 57 ABSTRACT 87, PCT Pub. No.: WO92/18632 A bacteriophage packaging site-based vector system is describe that involves cloning vectors and methods for their PCT Pub. Date: Oct. 29, 1992 use in preparing multiple copy number bacteriophage librar O ies of cloned DNA. The vector is based on a DNA segment Related U.S. Application Data that comprises a nucleotide sequence defining a bacterioph 63 abandoned.continuationin-part of ser, No. 685.215,y Apr 12, 1991, ligationage packaging means usedsite locatedto concatamerize between twofragments termining of DNA 6 having compatible ligation means. Methods for preparing a (51) Int. Cl. ............................... C12N 1/21; C12N 7/01; concatameric DNA for packaging cloned DNA segments, C12N 15/10; C12N 15/11 and for packaging the concatamers to produce a library are 52 U.S. Cl. ................ 435/914; 435/1723; 435/252.33; also described. 435/235.1; 435/252.3; 536/23.1 58) Field of Search .............................. 435/91.32, 172.3, 435/320.1,914, 252.33, 252.3, 235.1; 536/23.1 18 Claims, 1 Drawing Sheet U.S. -

Lessons from Nature: Microrna-Based Shrna Libraries
PERSPECTIVE Lessons from Nature: microRNA-based shRNA libraries methods Kenneth Chang1, Stephen J Elledge2 & Gregory J Hannon1 Loss-of-function genetics has proven essential for interrogating the functions of genes .com/nature e and for probing their roles within the complex circuitry of biological pathways. In .natur many systems, technologies allowing the use of such approaches were lacking before w the discovery of RNA interference (RNAi). We have constructed first-generation short hairpin RNA (shRNA) libraries modeled after precursor microRNAs (miRNAs) and second- http://ww generation libraries modeled after primary miRNA transcripts (the Hannon-Elledge oup libraries). These libraries were arrayed, sequence-verified, and cover a substantial portion r G of all known and predicted genes in the human and mouse genomes. Comparison of first- and second-generation libraries indicates that RNAi triggers that enter the RNAi pathway lishing through a more natural route yield more effective silencing. These large-scale resources b Pu are functionally versatile, as they can be used in transient and stable studies, and for constitutive or inducible silencing. Library cassettes can be easily shuttled into vectors Nature that contain different promoters and/or that provide different modes of viral delivery. 6 200 © RNAi is an evolutionarily conserved, sequence-specific than a decade elapsed between the discovery of the first gene-silencing mechanism that is induced by dsRNA. miRNA, Caenorhabditis elegans lin-4 (refs. 11,12) and Each dsRNA silencing trigger is processed into 21–25 bp the achievement of at least a superficial understanding dsRNAs called small interfering RNAs (siRNAs)1–3 by of the biosynthesis, processing and mode of action of Dicer, a ribonuclease III family (RNase III) enzyme4. -

Site Directed Mutagensis of Bacteriophage HK639 and Identification of Its Integration Site Madhuri Jonnalagadda Western Kentucky University
Western Kentucky University TopSCHOLAR® Masters Theses & Specialist Projects Graduate School 12-2008 Site Directed Mutagensis of Bacteriophage HK639 and Identification of Its Integration Site Madhuri Jonnalagadda Western Kentucky University Follow this and additional works at: http://digitalcommons.wku.edu/theses Part of the Cell and Developmental Biology Commons, Genetics Commons, Immunopathology Commons, and the Molecular Genetics Commons Recommended Citation Jonnalagadda, Madhuri, "Site Directed Mutagensis of Bacteriophage HK639 and Identification of Its Integration Site" (2008). Masters Theses & Specialist Projects. Paper 42. http://digitalcommons.wku.edu/theses/42 This Thesis is brought to you for free and open access by TopSCHOLAR®. It has been accepted for inclusion in Masters Theses & Specialist Projects by an authorized administrator of TopSCHOLAR®. For more information, please contact [email protected]. SITE DIRECTED MUTAGENESIS OF BACTERIOPHAGE HK639 AND IDENTIFICATION OF ITS INTEGRATION SITE A Thesis presented to The faculty of the Department of Biology Western Kentucky University Bowling Green, Kentucky. In Partial Fulfillment Of the Requirement for the Degree Master of Science By Madhuri Jonnalagadda December, 2008 Site Directed Mutagenesis of Bacteriophage HK639 and Identification of its Integration Site Date Recommended 12/4/08 Dr.Rodney A. King Director of Thesis __Dr.Jeffery Marcus Dr. Sigrid Jacobshagen _____________________________________ Dean, Graduate Studies and Research Date DEDICATION I would like to dedicate my thesis to my dear father Dr. J. Padmanabha Rao, my mother, Dr.V. Lakshmi, my sister Dr. J. Sushma for their support and encouragement. i ACKNOWLEDGEMENTS I wish to express my heartfelt gratitude to my graduate committee in the Department of Biology, Western Kentucky University. Primarily, I am thankful to my advisor, Dr. -

MORC1 Represses Transposable Elements in the Mouse Male Germline
ARTICLE Received 12 Sep 2014 | Accepted 7 Nov 2014 | Published 12 Dec 2014 DOI: 10.1038/ncomms6795 OPEN MORC1 represses transposable elements in the mouse male germline William A. Pastor1,*, Hume Stroud1,*,w, Kevin Nee1, Wanlu Liu1, Dubravka Pezic2, Sergei Manakov2, Serena A. Lee1, Guillaume Moissiard1, Natasha Zamudio3,De´borah Bourc’his3, Alexei A. Aravin2, Amander T. Clark1,4 & Steven E. Jacobsen1,4,5 The Microrchidia (Morc) family of GHKL ATPases are present in a wide variety of prokaryotic and eukaryotic organisms but are of largely unknown function. Genetic screens in Arabidopsis thaliana have identified Morc genes as important repressors of transposons and other DNA-methylated and silent genes. MORC1-deficient mice were previously found to display male-specific germ cell loss and infertility. Here we show that MORC1 is responsible for transposon repression in the male germline in a pattern that is similar to that observed for germ cells deficient for the DNA methyltransferase homologue DNMT3L. Morc1 mutants show highly localized defects in the establishment of DNA methylation at specific classes of transposons, and this is associated with failed transposon silencing at these sites. Our results identify MORC1 as an important new regulator of the epigenetic landscape of male germ cells during the period of global de novo methylation. 1 Department of Molecular, Cell and Developmental Biology, University of California Los Angeles, 4028 Terasaki Life Sciences Building, 610 Charles E. Young Drive East, Los Angeles, California 90095, USA. 2 Division of Biology and Biochemical Engineering, California Institute of Technology, 1200 E California Boulevard, Pasadena, California 91125, USA. 3 Unite´ de ge´ne´tique et biologie du de´veloppement, Instititute Curie, CNRS UMR3215, INSERM U934, Paris 75005, France. -

Herpesviral Latency—Common Themes
pathogens Review Herpesviral Latency—Common Themes Magdalena Weidner-Glunde * , Ewa Kruminis-Kaszkiel and Mamata Savanagouder Department of Reproductive Immunology and Pathology, Institute of Animal Reproduction and Food Research of Polish Academy of Sciences, Tuwima Str. 10, 10-748 Olsztyn, Poland; [email protected] (E.K.-K.); [email protected] (M.S.) * Correspondence: [email protected] Received: 22 January 2020; Accepted: 14 February 2020; Published: 15 February 2020 Abstract: Latency establishment is the hallmark feature of herpesviruses, a group of viruses, of which nine are known to infect humans. They have co-evolved alongside their hosts, and mastered manipulation of cellular pathways and tweaking various processes to their advantage. As a result, they are very well adapted to persistence. The members of the three subfamilies belonging to the family Herpesviridae differ with regard to cell tropism, target cells for the latent reservoir, and characteristics of the infection. The mechanisms governing the latent state also seem quite different. Our knowledge about latency is most complete for the gammaherpesviruses due to previously missing adequate latency models for the alpha and beta-herpesviruses. Nevertheless, with advances in cell biology and the availability of appropriate cell-culture and animal models, the common features of the latency in the different subfamilies began to emerge. Three criteria have been set forth to define latency and differentiate it from persistent or abortive infection: 1) persistence of the viral genome, 2) limited viral gene expression with no viral particle production, and 3) the ability to reactivate to a lytic cycle. This review discusses these criteria for each of the subfamilies and highlights the common strategies adopted by herpesviruses to establish latency. -

Bacterial Virus Ontology; Coordinating Across Databases
viruses Article Bacterial Virus Ontology; Coordinating across Databases Chantal Hulo 1, Patrick Masson 1, Ariane Toussaint 2, David Osumi-Sutherland 3, Edouard de Castro 1, Andrea H. Auchincloss 1, Sylvain Poux 1, Lydie Bougueleret 1, Ioannis Xenarios 1 and Philippe Le Mercier 1,* 1 Swiss-Prot group, SIB Swiss Institute of Bioinformatics, CMU, University of Geneva Medical School, 1211 Geneva, Switzerland; [email protected] (C.H.); [email protected] (P.M.); [email protected] (E.d.C.); [email protected] (A.H.A.); [email protected] (S.P.); [email protected] (L.B.); [email protected] (I.X.) 2 University Libre de Bruxelles, Génétique et Physiologie Bactérienne (LGPB), 12 rue des Professeurs Jeener et Brachet, 6041 Charleroi, Belgium; [email protected] 3 European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton CB10 1SD, UK; [email protected] * Correspondence: [email protected]; Tel.: +41-22379-5870 Academic Editors: Tessa E. F. Quax, Matthias G. Fischer and Laurent Debarbieux Received: 13 April 2017; Accepted: 17 May 2017; Published: 23 May 2017 Abstract: Bacterial viruses, also called bacteriophages, display a great genetic diversity and utilize unique processes for infecting and reproducing within a host cell. All these processes were investigated and indexed in the ViralZone knowledge base. To facilitate standardizing data, a simple ontology of viral life-cycle terms was developed to provide a common vocabulary for annotating data sets. New terminology was developed to address unique viral replication cycle processes, and existing terminology was modified and adapted. -
Virus Replication Cycles
© Jones and Bartlett Publishers. NOT FOR SALE OR DISTRIBUTION A scanning electron micrograph of Ebola virus particles. Ebola virus contains an RNA genome. It causes Ebola hemorrhagic fever, which is a severe and often fatal disease in hu- mans and nonhuman primates. CHAPTER Virus Replication Cycles OUTLINE 3.1 One-Step Growth Curves 3.3 The Error-Prone RNA Polymerases: 3 3.2 Key Steps of the Viral Replication Genetic Diversity Cycle 3.4 Targets for Antiviral Therapies In the struggle for survival, the ■ 1. Attachment (Adsorption) ■ RNA Virus Mutagens: A New Class “ ■ 2. Penetration (Entry) of Antiviral Drugs? fi ttest win out at the expense of ■ 3. Uncoating (Disassembly and Virus File 3-1: How Are Cellular Localization) their rivals because they succeed Receptors Used for Viral Attachment ■ 4. Types of Viral Genomes and Discovered? in adapting themselves best to Their Replication their environment. ■ 5. Assembly Refresher: Molecular Biology ” ■ 6. Maturation Charles Darwin ■ 7. Release 46 229329_CH03_046_069.indd9329_CH03_046_069.indd 4466 11/18/08/18/08 33:19:08:19:08 PPMM © Jones and Bartlett Publishers. NOT FOR SALE OR DISTRIBUTION CASE STUDY The campus day care was recently closed during the peak of the winter fl u season because many of the young children were sick with a lower respiratory tract infection. An email an- nouncement was sent to all students, faculty, and staff at the college that stated the closure was due to a metapneumovirus outbreak. The announcement briefed the campus com- munity with information about human metapneumonoviruses (hMPVs). The announcement stated that hMPV was a newly identifi ed respiratory tract pathogen discovered in the Netherlands in 2001. -

The Roles of Moron Genes in the Escherichia Coli Enterobacteria Phage Phi-80
THE ROLES OF MORON GENES IN THE ESCHERICHIA COLI ENTEROBACTERIA PHAGE PHI-80 Yury V. Ivanov A Dissertation Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY December 2012 Committee: Ray A. Larsen, Advisor Craig L. Zirbel Graduate Faculty Representative Vipa Phuntumart Scott O. Rogers George S. Bullerjahn © 2012 Yury Ivanov All Rights Reserved iii ABSTRACT Ray Larsen, Advisor The TonB system couples cytoplasmic membrane-derived proton motive force energy to drive ferric siderophore transport across the outer membrane of Gram-negative bacteria. While much effort has focused on this process, how energy is harnessed to provide for transport of ligands remains unknown. Several bacterial viruses (“phage”) are known to require the TonB system to irreversibly adsorb (i.e., establish infection) in the model organism Escherichia coli. One such phage is φ80, a “cousin” of the model temperate phage λ. Determining how φ80 is using the TonB system for infection should provide novel insights to the mechanisms of TonB-dependent processes. It had long been known that recombination between λ and φ80 results in a λ-like phage for whom TonB is now required; and this recombination involved the λ J gene, which encodes the tail-spike protein required for irreversible adsorption of λ to E. coli. Thus, we suspected that a φ80 homologue of the λ J gene product was responsible for the TonB dependence of φ80. While φ80 has long served as a tool for assaying TonB activity, it has not received the scrutiny afforded λ. -

Viruses Chapter 19
Viruses Chapter 19 What you must know: The components of a virus. The differences between lytic and lysogenic cycles. How viruses can introduce genetic variation into host organisms. Mechanisms that introduce genetic variation into viral populations. Bacteria vs. Viruses Bacteria Virus Prokaryotic cell Not a living cell (genes Most are free-living (some packaged in protein shell) parasitic) Intracellular parasite Relatively large size 1/1000 size of bacteria Antibiotics used to kill Vaccines used to prevent bacteria viral infection Antiviral treatment Viruses Very small (<ribosomes) Components = nucleic acid + capsid ◦ Nucleic acid: DNA or RNA (double or single-stranded) ◦ Capsid: protein shell ◦ Some viruses also have viral envelopes that surround capsid Derived from host cell membranes and viral proteins Viruses Limited host range ◦ Entry = attach to host cell membrane receptors through capsid proteins or glycoproteins on viral envelope (animal) ◦ Eg. human cold virus (rhinovirus) upper respiratory tract (mouth & nose) Reproduce quickly within host cells Can mutate easily ◦ RNA viruses: no error-checking mechanisms Simplified viral replicative cycle Entry: Injection endocytosis, fusion VIDEO: T4 PHAGE INFECTION Bacteriophage Virus that infects bacterial cells Viral Reproduction - phages Lytic Cycle: ◦ Use host machinery to replicate, assemble, and release copies of virus ◦ Virulent phages: Cells die through lysis or apoptosis Lysogenic (Latent) Cycle: ◦ DNA incorporated into host DNA and replicated along with it -

Ab Komplet 6.07.2018
CONTENTS 1. Welcome addresses 2 2. Introduction 3 3. Acknowledgements 10 4. General information 11 5. Scientific program 16 6. Abstracts – oral presentations 27 7. Abstracts – poster sessions 99 8. Participants 419 1 EMBO Workshop Viruses of Microbes 2018 09 – 13 July 2018 | Wrocław, Poland 1. WELCOME ADDRESSES Welcome to the Viruses of Microbes 2018 EMBO Workshop! We are happy to welcome you to Wrocław for the 5th meeting of the Viruses of Microbes series. This series was launched in the year 2010 in Paris, and was continued in Brussels (2012), Zurich (2014), and Liverpool (2016). This year our meeting is co-organized by two partner institutions: the University of Wrocław and the Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences. The conference venue (University of Wrocław, Uniwersytecka 7-10, Building D) is located in the heart of Wrocław, within the old, historic part of the city. This creates an opportunity to experience the over 1000-year history of the city, combined with its current positive energy. The Viruses of Microbes community is constantly growing. More and more researchers are joining it, and they represent more and more countries worldwide. Our goal for this meeting was to create a true global platform for networking and exchanging ideas. We are most happy to welcome representatives of so many countries and continents. To accommodate the diversity and expertise of the scientists and practitioners gathered by VoM2018, the leading theme of this conference is “Biodiversity and Future Application”. With the help of your contribution, this theme was developed into a program covering a wide range of topics with the strongest practical aspect.