Independently Evolving Species in Asexual Bdelloid Rotifers
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Rotifer Species Diversity in Mexico: an Updated Checklist
diversity Review Rotifer Species Diversity in Mexico: An Updated Checklist S. S. S. Sarma 1,* , Marco Antonio Jiménez-Santos 2 and S. Nandini 1 1 Laboratory of Aquatic Zoology, FES Iztacala, National Autonomous University of Mexico, Av. de Los Barrios No. 1, Tlalnepantla 54090, Mexico; [email protected] 2 Posgrado en Ciencias del Mar y Limnología, Universidad Nacional Autónoma de México, Ciudad Universitaria, Mexico City 04510, Mexico; [email protected] * Correspondence: [email protected]; Tel.: +52-55-56231256 Abstract: A review of the Mexican rotifer species diversity is presented here. To date, 402 species of rotifers have been recorded from Mexico, besides a few infraspecific taxa such as subspecies and varieties. The rotifers from Mexico represent 27 families and 75 genera. Molecular analysis showed about 20 cryptic taxa from species complexes. The genera Lecane, Trichocerca, Brachionus, Lepadella, Cephalodella, Keratella, Ptygura, and Notommata accounted for more than 50% of all species recorded from the Mexican territory. The diversity of rotifers from the different states of Mexico was highly heterogeneous. Only five federal entities (the State of Mexico, Michoacán, Veracruz, Mexico City, Aguascalientes, and Quintana Roo) had more than 100 species. Extrapolation of rotifer species recorded from Mexico indicated the possible occurrence of more than 600 species in Mexican water bodies, hence more sampling effort is needed. In the current review, we also comment on the importance of seasonal sampling in enhancing the species richness and detecting exotic rotifer taxa in Mexico. Keywords: rotifera; distribution; checklist; taxonomy Citation: Sarma, S.S.S.; Jiménez-Santos, M.A.; Nandini, S. Rotifer Species Diversity in Mexico: 1. -

Antarctic Bdelloid Rotifers: Diversity, Endemism and Evolution
1 Antarctic bdelloid rotifers: diversity, endemism and evolution 2 3 Introduction 4 5 Antarctica’s ecosystems are characterized by the challenges of extreme environmental 6 stresses, including low temperatures, desiccation and high levels of solar radiation, all of 7 which have led to the evolution and expression of well-developed stress tolerance features in 8 the native terrestrial biota (Convey, 1996; Peck et al., 2006). The availability of liquid water, 9 and its predictability, is considered to be the most important driver of biological and 10 biodiversity processes in the terrestrial environments of Antarctica (Block et al., 2009; 11 Convey et al., 2014). Antarctica’s extreme conditions and isolation combined with the over- 12 running of many, but importantly not all, terrestrial and freshwater habitats by ice during 13 glacial cycles, underlie the low overall levels of diversity that characterize the contemporary 14 faunal, floral and microbial communities of the continent (Convey, 2013). Nevertheless, in 15 recent years it has become increasingly clear that these communities contain many, if not a 16 majority, of species that have survived multiple glacial cycles over many millions of years 17 and undergone evolutionary radiation on the continent itself rather than recolonizing from 18 extra-continental refugia (Convey & Stevens, 2007; Convey et al., 2008; Fraser et al., 2014). 19 With this background, high levels of endemism characterize the majority of groups that 20 dominate the Antarctic terrestrial fauna, including in particular Acari, Collembola, Nematoda 21 and Tardigrada (Pugh & Convey, 2008; Convey et al., 2012). 22 The continent of Antarctica is ice-bound, and surrounded and isolated from the other 23 Southern Hemisphere landmasses by the vastness of the Southern Ocean. -

Rotifers: Excellent Subjects for the Study of Macro- and Microevolutionary Change
Hydrobiologia (2011) 662:11–18 DOI 10.1007/s10750-010-0515-1 ROTIFERA XII Rotifers: excellent subjects for the study of macro- and microevolutionary change Gregor F. Fussmann Published online: 1 November 2010 Ó Springer Science+Business Media B.V. 2010 Abstract Rotifers, both as individuals and as a nature. These features make them excellent eukaryotic phylogenetic group, are particularly worthwhile sub- model systems for the study of eco-evolutionary jects for the study of evolution. Over the past decade dynamics. molecular and experimental work on rotifers has facilitated major progress in three lines of evolution- Keywords Rotifer phylogeny Á Asexuality Á ary research. First, we continue to reveal the phy- Eco-evolutionary dynamics logentic relationships within the taxon Rotifera and its placement within the tree of life. Second, we have gained a better understanding of how macroevolu- Introduction tionary transitions occur and how evolutionary strat- egies can be maintained over millions of years. In the Rotifers are microscopic invertebrates that occur in case of rotifers, we are challenged to explain the freshwater, brackish water, in moss habitats, and in soil evolution of obligate asexuality (in the bdelloids) as (Wallace et al., 2006). Rotifers are particularly fasci- mode of reproduction and how speciation occurs in nating and suitable objects for the study of evolution the absence of sex. Recent research with bdelloid roti- and, over the past decade, featured prominently in fers has identified novel mechanisms such as horizon- research on both macro- and microevolutionary tal gene transfer and resistance to radiation as factors change. I, here, review the recent developments in potentially affecting macroevolutionary change. -

Thalassic Rotifers from the United States: Descriptions of Two New Species and Notes on the Effect of Salinity and Ecosystem on Biodiversity
diversity Article Thalassic Rotifers from the United States: Descriptions of Two New Species and Notes on the Effect of Salinity and Ecosystem on Biodiversity Francesca Leasi 1,* and Willem H. De Smet 2 1 Department of Biology, Geology and Environmental Science, University of Tennessee, Chattanooga, 615 McCallie Ave, Chattanooga, TN 37403, USA 2 Department of Biology. ECOBE, University of Antwerp, Universiteitsplein 1, 2610 Wilrijk, Belgium; [email protected] * Correspondence: [email protected] http://zoobank.org:pub:7679CE0E-11E8-4518-B132-7D23F08AC8FA Received: 26 November 2019; Accepted: 7 January 2020; Published: 13 January 2020 Abstract: This study shows the results of a rotifer faunistic survey in thalassic waters from 26 sites located in northeastern U.S. states and one in California. A total of 44 taxa belonging to 21 genera and 14 families were identified, in addition to a group of unidentifiable bdelloids. Of the fully identified species, 17 are the first thalassic records for the U.S., including Encentrum melonei sp. nov. and Synchaeta grossa sp. nov., which are new to science, and Colurella unicauda Eriksen, 1968, which is new to the Nearctic region. Moreover, a refined description of Encentrum rousseleti (Lie-Pettersen, 1905) is presented. During the survey, we characterized samples by different salinity values and ecosystems and compared species composition across communities to test for possible ecological correlations. Results indicate that both salinities and ecosystems are a significant predictor of rotifer diversity, supporting that biodiversity estimates of small species provide fundamental information for biomonitoring. Finally, we provide a comprehensive review of the diversity and distribution of thalassic rotifers in the United States. -

A Modern Approach to Rotiferan Phylogeny: Combining Morphological and Molecular Data
Molecular Phylogenetics and Evolution 40 (2006) 585–608 www.elsevier.com/locate/ympev A modern approach to rotiferan phylogeny: Combining morphological and molecular data Martin V. Sørensen ¤, Gonzalo Giribet Department of Organismic and Evolutionary Biology, Museum of Comparative Zoology, Harvard University, 16 Divinity Avenue, Cambridge, MA 02138, USA Received 30 November 2005; revised 6 March 2006; accepted 3 April 2006 Available online 6 April 2006 Abstract The phylogeny of selected members of the phylum Rotifera is examined based on analyses under parsimony direct optimization and Bayesian inference of phylogeny. Species of the higher metazoan lineages Acanthocephala, Micrognathozoa, Cycliophora, and potential outgroups are included to test rotiferan monophyly. The data include 74 morphological characters combined with DNA sequence data from four molecular loci, including the nuclear 18S rRNA, 28S rRNA, histone H3, and the mitochondrial cytochrome c oxidase subunit I. The combined molecular and total evidence analyses support the inclusion of Acanthocephala as a rotiferan ingroup, but do not sup- port the inclusion of Micrognathozoa and Cycliophora. Within Rotifera, the monophyletic Monogononta is sister group to a clade con- sisting of Acanthocephala, Seisonidea, and Bdelloidea—for which we propose the name Hemirotifera. We also formally propose the inclusion of Acanthocephala within Rotifera, but maintaining the name Rotifera for the new expanded phylum. Within Monogononta, Gnesiotrocha and Ploima are also supported by the data. The relationships within Ploima remain unstable to parameter variation or to the method of phylogeny reconstruction and poorly supported, and the analyses showed that monophyly was questionable for the fami- lies Dicranophoridae, Notommatidae, and Brachionidae, and for the genus Proales. -
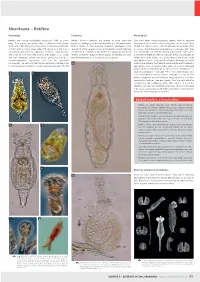
Microfauna – Rotifera
Microfauna – Rotifera Morphology Taxonomy Microhabitat Rotifers are minute multicellular organisms (0.05 to 3 mm Rotifers (phylum Rotifera) are related to other worm-like Like many other minute organisms, rotifers have an absolute long). Their mostly transparent body is subdivided into a head, organisms belonging to Gnathostomulida and Micrognathozoa. requirement for a water matrix during their active phase. They trunk, and a foot. They have three easily visible unique features: Recent studies in DNA evolution (molecular phylogeny) have inhabit the capillary water retained between soil particles, litter 1) their anterior ciliary organ called the corona (or crown); 2) a revealed that the parasitic worms of the phylum Acanthocephala or mosses, where they feed on bacteria or small algal cells. They specialised food processing apparatus made of strong muscles are their closest relatives, if not themselves a group of specialised are filter-feeders (i.e. feed by filtering food particles from water) and a set of hard jaws (the mastax with trophi); 3) a unique rotifers. Scientists recognise three groups of Rotifera, but only or browse the bacterium film for particles. A few are predators of and well developed cuticle (the lorica), giving the animals a one, the Bdelloidea, is an important soil inhabitant. ciliates or of other rotifers, or suck out the content of cells after pseudo-segmented appearance, that can be exquisitely piercing the cell wall using specialised trophi. Although they need ornamented. The head and foot can be retracted inside the trunk a water to live actively, the bdelloids, which are the most successful if the animal is disturbed or if the environment dries out. -

Sexual Reproduction in Bdelloid Rotifers
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.06.239590; this version posted August 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Sexual Reproduction in Bdelloid Rotifers Veronika N. Lainea, Timothy Sacktonb and Matthew Meselsonc aCurrent address, Department of Animal Ecology, Netherlands Institute of Ecology, 6708 PB Wageningen, The Netherlands; bInformatics Group, Faculty of Arts and Sciences, Harvard University, Cambridge, MA 02138; cDepartment of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138. Corresponding author: Email: [email protected] ABSTRACT Nearly all eukaryotes reproduce sexually, either constitutively or facultatively, and nearly all that are thought to be entirely asexual diverged recently from sexuals, implying that loss of sex leads to early extinction and therefore that sexual reproduction is essential for evolutionary success. Nevertheless, there are several groups that have been thought to have evolved asexually for millions of years. Of these, the most extensively studied are the rotifers of Class Bdelloidea. Yet the evidence for their asexuality is entirely negative -- the failure to establish the existence of males or hermaphrodites. Opposed to this is a growing body of evidence that bdelloids do reproduce sexually, albeit rarely, retaining meiosis-associated genes and, in a limited study of allele sharing among individuals of the bdelloid Macrotrachela quadricornifera, displaying a pattern of genetic exchange indicating recent sexual reproduction. Here we present a much larger study of allele sharing among these individuals clearly showing the occurrence of sexual reproduction, thereby removing what had appeared to be a serious challenge to hypotheses for the evolutionary advantage of sex and confirming that sexual reproduction is essential for long term evolutionary success in eukaryotes. -

Bipartition, Budding and Reproduction by Eggs
BioMEDIA ASSOCIATES LLC HIDDEN BIODIVERSITY Series Reproduction of microorganisms Study Guide Written and Photographed by Rubén Duro Pérez Supplement to Video Program All Text and Images ©2015 BioMEDIA ASSOCIATES LLC One of the characteristics of living beings is their ability to reproduce. All living beings reproduce, from the tiniest, such as bacteria, to the largest, such as trees or mammals. However, throughout evolution, each group of organisms has developed different strategies to perpetuate, and microscopic organisms have been no exception to this rule. BioMEDIA ASSOCIATES LLC • www.ebiomedia.com •[email protected] 877.661.5355 toll-free • 843.470.0236 voice • 843.470.0237 fax Page 1 The reproductive strategies adopted by microscopic organisms are varied. It is even possible to find organisms that do not have a single strategy but several, and use each depending on the environmental conditions. However, we could say that those found most frequently in this microscopic world are three: bipartition, budding and reproduction by eggs. Two of them, bipartition and budding, are asexual strategies, i.e. which does not involve sex, while the third, reproduction by eggs, may be sexual or asexual. Bipartition Bipartition is the strategy adopted by bacteria and protozoa. Organisms in which there are neither male nor female. And it is also used by cells that are part of our body. This process consists to divide by half the cell that forms the entire organism, so the result is the creation of two identical new organisms. During this division all the structures of the original cell are duplicated, and a complete pool of them goes to each of the new daughter cells. -

Epizoic and Parasitic Rotifers
Hydrobiologia 186/187: 59-67, 1989. C. Ricci, T. W. Snell and C. E. King (eds), Rotifer Symposium V. 59 © 1989 Kluwer Academic Publishers. Printed in Belgium. Epizoic and parasitic rotifers Linda May Institute of Freshwater Ecology, Edinburgh Research Station, Bush Estate, Penicuik, Midlothian EH26 OQB, Scotland, UK Key words: epizoic, parasitic, rotifers Abstract Many rotifer species live in close association with plants or other animals. Most of these associations are of a commensal or synoecious nature, some rotifer species having lost the ability to live independently. Few rotifers are true parasites, actually harming their hosts. The Seisonidae, Monogononta and Bdelloidea include epizoic and parasitic species. The most widely known are probably the parasites of colonial and filamentous algae (e.g. Volvox, Vaucheria). However, rotifers are also found on a wide range of invertebrates: colonial, sessile Protozoa; Porifera; Rotifera; Annelida; Bryozoa; Echinodermata; Mollusca, especially on the shells and egg masses of aquatic gastropods; Crustacea, including the lower forms (e.g. Daphnia, Asellus, Gammarus) and in the gill chambers of Astacus and Chasmagnathus; the aquatic larvae of insects. There appear to be few records of epizoic or parasitic rotifers among vertebrates, apart from Encentrum kozminskii on carp, Limnias ceratophylli on the Amazonian crocodile, Melanosuchus niger, and an unidentified Bdelloid apparently living as a pathogenic rotifer in Man. Introduction This paper reviews the literature which de- scribes parasitic or epizoic associations between Rotifers have long been known to live in close rotifers and other organisms. Much of this litera- association with other organisms, but little is ture was published in the late 1800's and early known of the types of relationships involved. -

Rotifera Bdelloidea
Rotifera Bdelloidea ROTIFERA BDELLOIDEA Diego Fontaneto Imperial College London, Division of Biology, Silwood Park Campus, Ascot Berkshire, SL57PY, United Kingdom [email protected] Abstract Bdelloid rotifers are microscopic aquatic animals, generally smaller than 1 mm. They can be found in any water body, but also in mosses, lichens and soil, due to their ability to survive desiccation entering dormancy. Dormant stages can also act as propagules for passive dispersal. These animals are very important in evolutionary biology, as they are the most diverse and old of the ‘ancient asexuals’. The following text is adapted from Fontaneto, D. & Ricci, C. (2004) Rotifera: Bdelloidea. In: Yule C.M. & Yong H.S. (eds.), Freshwater invertebrates of the Malaysian Region. Academy of Sciences Malaysia, Kuala Lumpur, Malaysia, pp. 121-126. Additional information on rotifers in general can be found in Wallace R.L., Snell T.W., Ricci C. & Nogrady T., 2006. Rotifera. Volume 1: Biology, Ecology and Systematics (2nd edition). In: Dumont H.J.F., Guides to the identification of the microinvertebrates of the world. Kenobi Productions and Backhuys Publishers. 299 pp. An online key for families and genera of monogonont rotifers, limited to the marine habitat is available from http://www.pfeil-verlag.de/04biol/pdf/mm16_03.pdf Summer School in Taxonomy, Valdieri, Italy page 1 of 11 Rotifera Bdelloidea 1. Introduction The Bdelloidea is a class of rotifers that reproduce through ameiotic parthenogenesis only, and that are very common in several aquatic habitats (Donner 1965; Gilbert 1983; Ricci 1992; Mark Welch and Meselson 2000), and they have been named “evolutionary scandals” by Maynard Smith (1986), as they survived and speciated in the absence of sex. -

Chapter 10. Asexual Speciation
1 Lost Sex 2 The Evolutionary Biology of Parthenogenesis 3 Edited by Peter Van Dijk, Koen Martens, Isa Schön 4 5 Chapter 10. Asexual Speciation 6 C. William Birky, Jr. 7 Department of Ecology and Evolutionary Biology, The University of Arizona, Tucson, AZ 8 85745 USA 9 Phone 520-626-6513 10 Fax 520-621-9190 11 Email [email protected] 12 13 Timothy G. Barraclough 14 Division of Biology, Imperial College London, Silwood Park Campus, Ascot, Berkshire 15 SL5 7PY, UK 16 The Jodrell Laboratory, Royal Botanic Gardens, Kew TW9 3DS, UK 17 18 Phone: +44 (0)20 7594 2247 19 Fax: 44 (0)20 7594 2339 20 Email: [email protected] 21 22 The Importance of Asexual Species 23 24 There is a general consensus among biologists that species are real and important units of 25 biological diversity, and understanding the mechanisms of speciation is a hot research 26 topic (Coyne and Orr 2004). Nearly all of that work is focused on species and speciation 27 in animals and plants that reproduce sexually. But it is also critically important to 28 understand species and their origins in asexual organisms such as parthenogenetic 29 animals and plants. A substantial part of the earth’s biological diversity consists of 30 organisms that reproduce only asexually so far as we know, or reproduce sexually very 31 rarely or with extreme inbreeding so that they are effectively asexual or clonal. Asexual 32 organisms include not only the parthenogenetic animals and plants that are the subject of 33 this book, but also many fungi; some green algae, including the large and ancient genus 34 Chlorella; and many other eukaryotic protists, including possibly all of the members of 35 one of the five major groups of eukaryotes, the Excavates. -

Uzbekistan on the Conservation of Biological Diversity
THE SIXTH NATIONAL REPORT OF THE REPUBLIC OF UZBEKISTAN ON THE CONSERVATION OF BIOLOGICAL DIVERSITY THE UNITED NATIONS DEVELOPMENT PROGRAMME IN UZBEKISTAN GLOBAL ENVIRONMENT FACILITY STATE COMMITTEE OF THE REPUBLIC OF UZBEKISTAN ON ECOLOGY AND ENVIRONMENTAL PROTECTION THE SIXTH NATIONAL REPORT OF THE REPUBLIC OF UZBEKISTAN ON THE CONSERVATION OF BIOLOGICAL DIVERSITY Tashkent 2018 2 THE SIXTH NATIONAL REPORT OF THE REPUBLIC OF UZBEKISTAN ON THE CONSERVATION OF BIOLOGICAL DIVERSITY Sixth National Report of the Republic of Uzbekistan on the conservation of biological diversity / edited by B.T. Kuchkarov / Tashkent, 2018. - 207p. The report was prepared under the overall guidance of B.T. Kuchkarov, the Chairperson of the State Committee of the Republic of Uzbekistan on Ecology and Environmental Protection (Goskomekologiya RUz) and National Coordinator of the Project “Technical Support to Eligible Parties to Produce the Sixth National Report to the Convention on Biological Diversity”. Authors: Kh. Sherimbetov, Candidate of Technical Sciences, Head of the Department on Protected Natural Areas, the State Committee for Ecology and Environmental Protection of the Republic of Uzbekistan, Team Leader M. Aripdjanov, a.i., Head of the Bioinspection, the State Committee for Ecology and Environmental Protection of the Republic of Uzbekistan R. Gabitova, Gender Specialist Y. Mitropolskaya, Candidate of Biological Sciences, Senior Scientist, Institute of Zoology, Academy of Sciences of Uzbekistan U. Sobirov, Head of the Department for Biodiversity and Protected Natural Areas, the State Committee for Ecology and Environmental Protection of the Republic of Uzbekistan V. Talskikh, Candidate of Biological Sciences, Head of the Information Department of the Environmental Pollution Monitoring Service, Uzhydromet at Ministry of Emergency Situations of the Republic of Uzbekistan O.