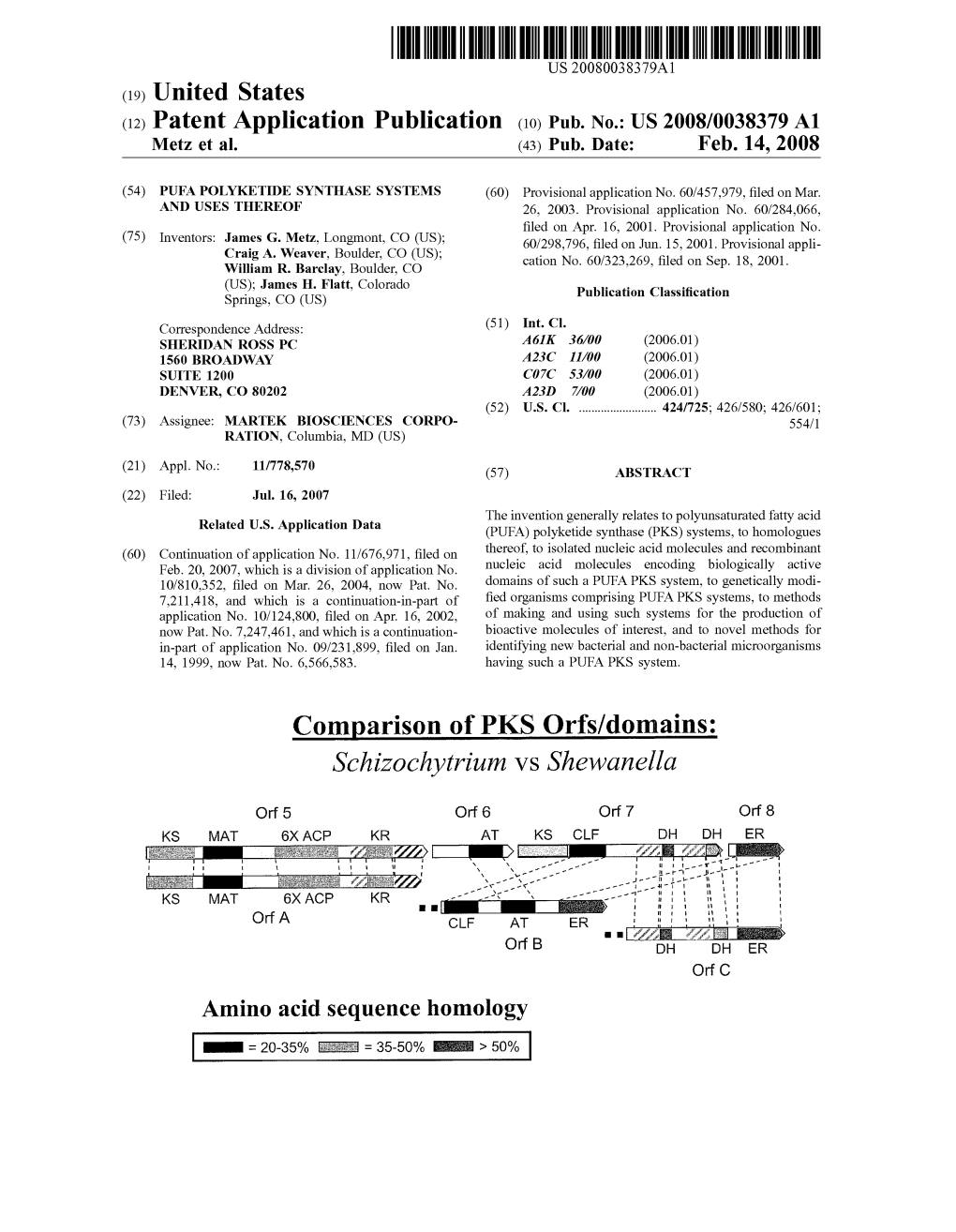
Schizochytrium VS Shewanella
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Physiology and Biochemistry of the Tropical Seagrass Thalassia Testudinum in Response to Hypersalinity Stress and Labyrinthula Sp
UNF Digital Commons UNF Graduate Theses and Dissertations Student Scholarship 2011 Physiology and Biochemistry of the Tropical Seagrass Thalassia testudinum in Response to Hypersalinity Stress and Labyrinthula sp. Infection Stacey Marie Trevathan-Tackett University of North Florida Suggested Citation Trevathan-Tackett, Stacey Marie, "Physiology and Biochemistry of the Tropical Seagrass Thalassia testudinum in Response to Hypersalinity Stress and Labyrinthula sp. Infection" (2011). UNF Graduate Theses and Dissertations. 391. https://digitalcommons.unf.edu/etd/391 This Master's Thesis is brought to you for free and open access by the Student Scholarship at UNF Digital Commons. It has been accepted for inclusion in UNF Graduate Theses and Dissertations by an authorized administrator of UNF Digital Commons. For more information, please contact Digital Projects. © 2011 All Rights Reserved Physiology and Biochemistry of the Tropical Seagrass Thalassia testudinum in Response to Hypersalinity Stress and Labyrinthula sp. Infection by Stacey Marie Trevathan-Tackett A thesis submitted to the Department of Biology in partial fulfillment of the requirements for the degree of Master of Science in Biology University of North Florida College of Arts and Sciences December, 2011 Unpublished Work © 2011 Stacey Marie Trevathan-Tackett Signature Deleted Signature Deleted Signature Deleted Signature Deleted Signature Deleted Signature Deleted Acknowledgements I first would like to acknowledge NOAA and the Nancy Foster Scholarship for funding both school and living expenses for the majority of my Master‟s career allowing me to focus all of my time and efforts toward research. Also, I would like to thank the Dodson Grant, the Coastal Biology program and the Graduate School at UNF for their continued financial support and resources during graduate school. -

Genetic Diversity of Labyrinthula Spp. in Seagrass Zostera Marina Beds in the North Atlantic Ocean and the Baltic Sea
Genetic diversity of Labyrinthula spp. in seagrass Zostera marina beds in the North Atlantic Ocean and the Baltic Sea Submitted by Matsapume Detcharoen In partial fulfillment of the requirements for the Degree of Master of Science in Biology Ludwig-Maximilians-Universität München Faculty of Biology August 2015 Abstract Die sogenannte Seegras – „Wasting Disease“ führte in den 1930er Jahren zu einem Massenrückgang des Seegrases Zostera marina im nördlichen Atlantik. Labyrinthula zosterae ist der potentielle Erreger der Krankheit, der zu dem Verlust von Seegrasbetten führen kann. Dieser Organismus infiziert die Blätter des Seegrases. Frühere Studien, die die Vielfalt der Labyrinthula spp untersuchten, basierten auf der Kultivierung von Labyrinthula. Diese Studie ist die erste Studie, die Blattproben zur molekularen Identifizierung von Labyrinthula verwendet und erforscht die Vielfalt der Gattung Labyrinthula. Proben von sechs Standorten wurden untersucht, dabei wurde ein 290 bp lange Teilsequenz der 18S rDNA kloniert und Sanger sequenziert. Das Abgleichen der erhaltenen Sequenzen mit NCBI GenBank mittels des blast Algorithmus ergab, dass die meisten Sequenzen von 77 bis 100% mit L. zosterae übereinstimmen, während inige Sequenzen der Standorte Wackerballig und Sandspollen eine 84-94 prozentige Übereinstimmung mit Labyrinthula spp zeigten. Keine der mutmaßlichen Labyrinthula spp. Sequenzen wurden mit bekannten Sequenzen aus der Datenbank zusammengefasst. Es wurden sieben OTUs generiert. Der Test von verschiedenen DNA-Extraktionmethoden ergab für ein Pflanzen- und ein Gewebe-Extraktionskit ähnliche Ergebnisse. Nichtsdestotrotz hat das Pflanzen-Kit mehr Vorteile wegen geringerer Kosten und dem geringeren Zeitaufwand. Zusammenfassend zeigt diese Studie, dass bisher unbekannte Labyrinthula spp. in den Seegraspopulationen vorkommen, und dass das in dieser Studie verwendete Verfahren für weitere Anwendungen geeignet ist. -

Associated Heterotrophic Protists, Stereomyxa Ramosa, Nitzschia Alba and Labyrinthula Sp
AQUATIC MICROBIAL ECOLOGY Vol. 21: 49-57,2000 Published February 21 Aquat Microb Ecol Utilisation of seaweed carbon by three surface- associated heterotrophic protists, Stereomyxa ramosa, Nitzschia alba and Labyrinthula sp. Evelyn ~rmstrong'~~~*,Andrew ~ogerson~,John W. ~eftley~ 'University Marine Biological Station Millport, Isle of Curnbrae KA28 OEG, Scotland, UK 'Oceanographic Center, Nova Southeastern University, 8000 N. Ocean Drive, Dania Beach, Florida 33004, USA 'Scottish Association for Marine Science, Dunstaffnage Marine Laboratory. PO Box 3, Oban. Argyll PA34 4AD. Scotland, UK ABSTRACT: In view of the abundance of protists associated with seaweeds and the diversity of nutri- tional strategies displayed by protists in general, the ability of 3 closely associated protists to utilise sea- weed carbon was investigated. Stereomyxa ramosa, Nitzschja alba and Labyrinthula sp. were cultured with seaweed polysaccharides as well as seaweed itself. N. alba and Labyrinthula sp. were found to utilise seaweed polysaccharides in axenic culture. All 3 protists were capable of penetrating intact but 'damaged' (autoclaved) seaweed particularly when bacteria were present. The possibility that these and other heterotrophic protists are directly removing macroalgal carbon in the field is discussed. KEY WORDS: Protist . Heterotroph - Seaweed. Carbon . Utilisation INTRODUCTION (Lucas et al. 1981, Rieper-Kirchner 1989).This conver- sion of dissolved carbon into particulate forms (as bac- Heterotrophic protists are abundant on seaweed sur- terial biomass) is thought to be a major link to higher faces (Rogerson 1991, Armstrong et al. 2000) because trophic levels (Newell et al. 1980). To compete effec- seaweeds are rich in bacterial prey (Laycock 1974, tively with bacteria, heterotrophic protists would need Shiba & Taga 1980). -

Bacterial Community Structure in a Sympagic Habitat Expanding with Global Warming: Brackish Ice Brine at 85€“90 °N
The ISME Journal (2019) 13:316–333 https://doi.org/10.1038/s41396-018-0268-9 ARTICLE Bacterial community structure in a sympagic habitat expanding with global warming: brackish ice brine at 85–90 °N 1,11 1,2 3 4 5,8 Beatriz Fernández-Gómez ● Beatriz Díez ● Martin F. Polz ● José Ignacio Arroyo ● Fernando D. Alfaro ● 1 1 2,6 5 4,7 Germán Marchandon ● Cynthia Sanhueza ● Laura Farías ● Nicole Trefault ● Pablo A. Marquet ● 8,9 10 10 Marco A. Molina-Montenegro ● Peter Sylvander ● Pauline Snoeijs-Leijonmalm Received: 12 February 2018 / Revised: 11 June 2018 / Accepted: 24 July 2018 / Published online: 18 September 2018 © International Society for Microbial Ecology 2018 Abstract Larger volumes of sea ice have been thawing in the Central Arctic Ocean (CAO) during the last decades than during the past 800,000 years. Brackish brine (fed by meltwater inside the ice) is an expanding sympagic habitat in summer all over the CAO. We report for the first time the structure of bacterial communities in this brine. They are composed of psychrophilic extremophiles, many of them related to phylotypes known from Arctic and Antarctic regions. Community structure displayed strong habitat segregation between brackish ice brine (IB; salinity 2.4–9.6) and immediate sub-ice seawater (SW; – 1234567890();,: 1234567890();,: salinity 33.3 34.9), expressed at all taxonomic levels (class to genus), by dominant phylotypes as well as by the rare biosphere, and with specialists dominating IB and generalists SW. The dominant phylotypes in IB were related to Candidatus Aquiluna and Flavobacterium, those in SW to Balneatrix and ZD0405, and those shared between the habitats to Halomonas, Polaribacter and Shewanella. -

Controlled Sampling of Ribosomally Active Protistan Diversity in Sediment-Surface Layers Identifies Putative Players in the Marine Carbon Sink
The ISME Journal (2020) 14:984–998 https://doi.org/10.1038/s41396-019-0581-y ARTICLE Controlled sampling of ribosomally active protistan diversity in sediment-surface layers identifies putative players in the marine carbon sink 1,2 1 1 3 3 Raquel Rodríguez-Martínez ● Guy Leonard ● David S. Milner ● Sebastian Sudek ● Mike Conway ● 1 1 4,5 6 7 Karen Moore ● Theresa Hudson ● Frédéric Mahé ● Patrick J. Keeling ● Alyson E. Santoro ● 3,8 1,9 Alexandra Z. Worden ● Thomas A. Richards Received: 6 October 2019 / Revised: 4 December 2019 / Accepted: 17 December 2019 / Published online: 9 January 2020 © The Author(s) 2020. This article is published with open access Abstract Marine sediments are one of the largest carbon reservoir on Earth, yet the microbial communities, especially the eukaryotes, that drive these ecosystems are poorly characterised. Here, we report implementation of a sampling system that enables injection of reagents into sediments at depth, allowing for preservation of RNA in situ. Using the RNA templates recovered, we investigate the ‘ribosomally active’ eukaryotic diversity present in sediments close to the water/sediment interface. We 1234567890();,: 1234567890();,: demonstrate that in situ preservation leads to recovery of a significantly altered community profile. Using SSU rRNA amplicon sequencing, we investigated the community structure in these environments, demonstrating a wide diversity and high relative abundance of stramenopiles and alveolates, specifically: Bacillariophyta (diatoms), labyrinthulomycetes and ciliates. The identification of abundant diatom rRNA molecules is consistent with microscopy-based studies, but demonstrates that these algae can also be exported to the sediment as active cells as opposed to dead forms. -

The Classification of Lower Organisms
The Classification of Lower Organisms Ernst Hkinrich Haickei, in 1874 From Rolschc (1906). By permission of Macrae Smith Company. C f3 The Classification of LOWER ORGANISMS By HERBERT FAULKNER COPELAND \ PACIFIC ^.,^,kfi^..^ BOOKS PALO ALTO, CALIFORNIA Copyright 1956 by Herbert F. Copeland Library of Congress Catalog Card Number 56-7944 Published by PACIFIC BOOKS Palo Alto, California Printed and bound in the United States of America CONTENTS Chapter Page I. Introduction 1 II. An Essay on Nomenclature 6 III. Kingdom Mychota 12 Phylum Archezoa 17 Class 1. Schizophyta 18 Order 1. Schizosporea 18 Order 2. Actinomycetalea 24 Order 3. Caulobacterialea 25 Class 2. Myxoschizomycetes 27 Order 1. Myxobactralea 27 Order 2. Spirochaetalea 28 Class 3. Archiplastidea 29 Order 1. Rhodobacteria 31 Order 2. Sphaerotilalea 33 Order 3. Coccogonea 33 Order 4. Gloiophycea 33 IV. Kingdom Protoctista 37 V. Phylum Rhodophyta 40 Class 1. Bangialea 41 Order Bangiacea 41 Class 2. Heterocarpea 44 Order 1. Cryptospermea 47 Order 2. Sphaerococcoidea 47 Order 3. Gelidialea 49 Order 4. Furccllariea 50 Order 5. Coeloblastea 51 Order 6. Floridea 51 VI. Phylum Phaeophyta 53 Class 1. Heterokonta 55 Order 1. Ochromonadalea 57 Order 2. Silicoflagellata 61 Order 3. Vaucheriacea 63 Order 4. Choanoflagellata 67 Order 5. Hyphochytrialea 69 Class 2. Bacillariacea 69 Order 1. Disciformia 73 Order 2. Diatomea 74 Class 3. Oomycetes 76 Order 1. Saprolegnina 77 Order 2. Peronosporina 80 Order 3. Lagenidialea 81 Class 4. Melanophycea 82 Order 1 . Phaeozoosporea 86 Order 2. Sphacelarialea 86 Order 3. Dictyotea 86 Order 4. Sporochnoidea 87 V ly Chapter Page Orders. Cutlerialea 88 Order 6. -

Short Communication on the Classification of The
SHORT COMMUNICATION ON THE CLASSIFICATION OF THE GENERA Labyrinthula, Schizochytrium AND Thraustochytrium (Labyrinthulids AND Thraustochytrids) Øjvind Moestrup Biological Institute, University of Copenhagen Universitetsparken 4 , DK- 2100 Copenhagen, Denmark E-mail: [email protected] Citation as: Moestrup Øjvind, 2019. On the classification of the genera Labyrinthula, Schizochytrium and Thraustochytrium (Labyrinthulids and Thraustochytrids). Tap chi Sinh hoc, 41(2): xx–xx. https://doi.org/10.15625/0866-7160/v41n2.xxxxx Species of the genera Labyrinthula, Schizochytrium, Thraustochytrium and related organisms have recently attracted attention in biotechnology, and here is a short note on how to classify these rather special organisms. The labyrinthulids and thraustochytrids belong to the heterokonts, a large group of very diverse organisms, from microscopic unicells to metre-long brown algae. The heterokonts comprise species that were formerly classified as algae, fungi and/or protozoa. Many heterokonts are autotrophic and contain chloroplasts, and such organisms are often classified as algae (golden algae, brown algae). Labyrinthulids and thraustochytrids, however, are heterotrophic and lack chloroplasts. They were until recently known as Labyrinthulomycetes or Labyrinthulea , indeed the new classification of Adl et al. (2019) uses the first of these names. It is an unfortunate name as it gives the misleading impression that they are fungi. Honigberg et al. (1964) and others considered them protozoa. Are heterokonts algae, protozoa or fungi? And, more specifically, what are labyrinthulids and thraustochytrids? The heterokonts are thought to be a very old group (probably precambrian), and this may account for their huge morphological diversity. In the WORMS list (World Register of Marine Species) heterokonts are classified as the Infrakingdom Heterokonta . -

Marine Phytophthora Species Can Hamper Conservation and Restoration of Vegetated Coastal Ecosystems
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by College of William & Mary: W&M Publish W&M ScholarWorks VIMS Articles Virginia Institute of Marine Science 2016 Marine Phytophthora species can hamper conservation and restoration of vegetated coastal ecosystems LL Govers WAM in 't Veid JP Meffert TJ Bouma PCJ van Rijswick See next page for additional authors Follow this and additional works at: https://scholarworks.wm.edu/vimsarticles Part of the Aquaculture and Fisheries Commons Recommended Citation Govers, LL; in 't Veid, WAM; Meffert, JP; Bouma, TJ; van Rijswick, PCJ; Heusinkveld, JHT; Orth, R J.; van Katwijk, MM; and van der Heide, T, "Marine Phytophthora species can hamper conservation and restoration of vegetated coastal ecosystems" (2016). VIMS Articles. 795. https://scholarworks.wm.edu/vimsarticles/795 This Article is brought to you for free and open access by the Virginia Institute of Marine Science at W&M ScholarWorks. It has been accepted for inclusion in VIMS Articles by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. Authors LL Govers, WAM in 't Veid, JP Meffert, TJ Bouma, PCJ van Rijswick, JHT Heusinkveld, R J. Orth, MM van Katwijk, and T van der Heide This article is available at W&M ScholarWorks: https://scholarworks.wm.edu/vimsarticles/795 Downloaded from http://rspb.royalsocietypublishing.org/ on October 25, 2018 Marine Phytophthora species can hamper rspb.royalsocietypublishing.org conservation and restoration of vegetated coastal ecosystems Laura L. Govers1,3, Willem A. Man in ‘t Veld4, Johan P. Meffert4, Tjeerd Research J. -

Draft Genomic Sequence of a Chromate- and Sulfate-Reducing
Xia et al. Standards in Genomic Sciences (2016) 11:48 DOI 10.1186/s40793-016-0169-3 SHORT GENOME REPORT Open Access Draft genomic sequence of a chromate- and sulfate-reducing Alishewanella strain with the ability to bioremediate Cr and Cd contamination Xian Xia1, Jiahong Li1, Shuijiao Liao1,2, Gaoting Zhou1,2, Hui Wang1, Liqiong Li1, Biao Xu1 and Gejiao Wang1* Abstract Alishewanella sp. WH16-1 (= CCTCC M201507) is a facultative anaerobic, motile, Gram-negative, rod-shaped bacterium isolated from soil of a copper and iron mine. This strain efficiently reduces chromate (Cr6+) to the much 3+ 2− 2− 2− 2+ less toxic Cr . In addition, it reduces sulfate (SO4 )toS . The S could react with Cd to generate precipitated CdS. Thus, strain WH16-1 shows a great potential to bioremediate Cr and Cd contaimination. Here we describe the features of this organism, together with the draft genome and comparative genomic results among strain WH16-1 and other Alishewanella strains. The genome comprises 3,488,867 bp, 50.4 % G + C content, 3,132 protein-coding genes and 80 RNA genes. Both putative chromate- and sulfate-reducing genes are identified. Keywords: Alishewanella, Chromate-reducing bacterium, Sulfate-reducing bacterium, Cadmium, Chromium Abbreviation: PGAP, Prokaryotic Genome Annotation Pipeline Introduction Alishewanella sp. WH16-1 was isolated from mining The genus Alishewanella was established by Vogel et al., soil in 2009. This strain could resist to multiple heavy in 2000 with Alishewanella fetalis as the type species. It metals. During cultivation, it could efficiently reduce the belongs to the family Alteromonadaceae of the class toxic chromate (Cr6+) to the much less toxic and less 3+ 2− Gammaproteobacteria [1]. -

Shewanella Oneidensis
RESEARCH ARTICLE Genome sequence of the dissimilatory metal ion–reducing bacterium Shewanella oneidensis John F.Heidelberg1,2, Ian T. Paulsen1,3, Karen E. Nelson1, Eric J. Gaidos4,5, William C. Nelson1, Timothy D. Read1, Jonathan A. Eisen1,3, Rekha Seshadri1, Naomi Ward1,2, Barbara Methe1, Rebecca A. Clayton1, Terry Meyer6, Alexandre Tsapin4, James Scott7, Maureen Beanan1, Lauren Brinkac1, Sean Daugherty1, Robert T. DeBoy1, Robert J. Dodson1, A. Scott Durkin1, Daniel H. Haft1, James F.Kolonay1, Ramana Madupu1, Jeremy D. Peterson1, Lowell A. Umayam1, Owen White1, Alex M. Wolf1, Jessica Vamathevan1, Janice Weidman1, Marjorie Impraim1, Kathy Lee1, Kristy Berry1, Chris Lee1, Jacob Mueller1, Hoda Khouri1, John Gill1, Terry R. Utterback1, Lisa A. McDonald1, Tamara V. Feldblyum1, Hamilton O. Smith1,8, J. Craig Venter1,9, Kenneth H. Nealson4,10, and Claire M. Fraser1,11* Published online 7 October 2002; doi:10.1038/nbt749 Shewanella oneidensis is an important model organism for bioremediation studies because of its diverse res- piratory capabilities, conferred in part by multicomponent, branched electron transport systems. Here we report the sequencing of the S. oneidensis genome, which consists of a 4,969,803–base pair circular chromo- http://www.nature.com/naturebiotechnology some with 4,758 predicted protein-encoding open reading frames (CDS) and a 161,613–base pair plasmid with 173 CDSs. We identified the first Shewanella lambda-like phage, providing a potential tool for further genome engineering. Genome analysis revealed 39 c-type cytochromes, including 32 previously unidentified in S. oneidensis, and a novel periplasmic [Fe] hydrogenase, which are integral members of the electron trans- port system. -

To Multi-Cell Level Charge Transport in Geobacter Sulfurreducens DL-1
ARTICLE Received 10 May 2013 | Accepted 10 Oct 2013 | Published 8 Nov 2013 DOI: 10.1038/ncomms3751 Probing single- to multi-cell level charge transport in Geobacter sulfurreducens DL-1 Xiaocheng Jiang1,*, Jinsong Hu2,*, Emily R. Petersen3, Lisa A. Fitzgerald4, Charles S. Jackan1, Alexander M. Lieber1, Bradley R. Ringeisen4, Charles M. Lieber1,5 & Justin C. Biffinger4 Microbial fuel cells, in which living microorganisms convert chemical energy into electricity, represent a potentially sustainable energy technology for the future. Here we report the single-bacterium level current measurements of Geobacter sulfurreducens DL-1 to elucidate the fundamental limits and factors determining maximum power output from a microbial fuel cell. Quantized stepwise current outputs of 92(±33) and 196(±20) fA are generated from microelectrode arrays confined in isolated wells. Simultaneous cell imaging/tracking and current recording reveals that the current steps are directly correlated with the contact of one or two cells with the electrodes. This work establishes the amount of current generated by an individual Geobacter cell in the absence of a biofilm and highlights the potential upper limit of microbial fuel cell performance for Geobacter in thin biofilms. 1 Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts 02138, USA. 2 CAS Key Laboratory of Molecular Nanostructure and Nanotechnology, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China. 3 Nova Research, Inc., 1900 Elkin Street, Suite 230, Alexandria, Virginia 22308, USA. 4 Chemistry Division, US Naval Research Laboratory, 4555 Overlook Avenue, SW, Washington, District of Columbia 20375, USA. 5 School of Engineering and Applied Science, Harvard University, Cambridge, Massachusetts 02138, USA. -

Seagrass Communities of the Gulf Coast of Florida: Status and Ecology
CLINTON J. DAWES August 2004 RONALD C. PHILLIPS GEROLD MORRISON CLINTON J. DAWES University of South Florida Tampa, Florida, USA RONALD C. PHILLIPS Institute of Biology of the Southern Seas Sevastopol, Crimea, Ukraine GEROLD MORRISON Environmental Protection Commission of Hillsborough County Tampa, Florida, USA August 2004 COPIES This document may be obtained from the following agencies: Tampa Bay Estuary Program FWC Fish and Wildlife Research Institute 100 8th Avenue SE 100 8th Avenue SE Mail Station I-1/NEP ATTN: Librarian St. Petersburg, FL 33701-5020 St. Petersburg, FL 33701-5020 Tel 727-893-2765 Fax 727-893-2767 Tel 727-896-8626 Fax 727-823-0166 www.tbep.org http://research.MyFWC.com CITATION Dawes, C.J., R.C. Phillips, and G. Morrison. 2004. Seagrass Communities of the Gulf Coast of Florida: Status and Ecology. Florida Fish and Wildlife Conservation Commission Fish and Wildlife Research Institute and the Tampa Bay Estuary Program. St. Petersburg, FL. iv + 74 pp. AUTHORS Clinton J. Dawes, Ph.D. Distinguished University Research Professor University of South Florida Department of Biology Tampa, FL 33620 [email protected] Ronald C. Phillips, Ph.D. Associate Institute of Biology of the Southern Seas 2, Nakhimov Ave. Sevastopol 99011 Crimea, Ukraine [email protected] Gerold Morrison, Ph.D. Director, Environmental Resource Management Environmental Protection Commission of Hillsborough County 3629 Queen Palm Drive Tampa, FL 33619 813-272-5960 ext 1025 [email protected] ii TABLE of CONTENTS iv Foreword and Acknowledgements 1 Introduction 6 Distribution, Status, and Trends 15 Autecology and Population Genetics 28 Ecological Roles 42 Natural and Anthropogenic Effects 49 Appendix: Taxonomy of Florida Seagrasses 55 References iii FOREWORD The waters along Florida’s Gulf of Mexico coastline, which stretches from the tropical Florida Keys in the south to the temperate Panhandle in the north, contain the most extensive and diverse seagrass meadows in the United States.