CRISPR-Cas9 Protein for Genome Editing
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Improving Treatment of Genetic Diseases with Crispr-Cas9 Rna-Guided Genome Editing
Sanchez 3:00 Team R06 Disclaimer: This paper partially fulfills a writing requirement for first-year (freshmen) engineering students at the University of Pittsburgh Swanson School of Engineering. This paper is a student paper, not a professional paper. This paper is not intended for publication or public circulation. This paper is based on publicly available information, and while this paper might contain the names of actual companies, products, and people, it cannot and does not contain all relevant information/data or analyses related to companies, products, and people named. All conclusions drawn by the authors are the opinions of the authors, first- year (freshmen) students completing this paper to fulfill a university writing requirement. If this paper or the information therein is used for any purpose other than the authors' partial fulfillment of a writing requirement for first-year (freshmen) engineering students at the University of Pittsburgh Swanson School of Engineering, the users are doing so at their own--not at the students', at the Swanson School's, or at the University of Pittsburgh's--risk. IMPROVING TREATMENT OF GENETIC DISEASES WITH CRISPR-CAS9 RNA-GUIDED GENOME EDITING Arijit Dutta [email protected] , Benjamin Ahlmark [email protected], Nate Majer [email protected] Abstract—Genetic illnesses are among the most difficult to treat as it is challenging to safely and effectively alter DNA. INTRODUCTION: THE WHAT, WHY, AND DNA is the basic code for all hereditary traits, so any HOW OF CRISPR-CAS9 alteration to DNA risks fundamentally altering the way someone’s genes are expressed. This change could lead to What Is CRISPR-Cas9? unintended consequences for both the individual whose DNA was altered and any offspring they may have in the future, CRISPR-Cas9 is an acronym that stands for “Clustered compounding the risk. -

CRISPR/Cas9: Tools and Applications for Eukaryotic Genome Editing
CRISPR/Cas9: Tools and Applications for Eukaryotic Genome Editing Fei Ann Ran Broad Institute Cambridge, Massachusetts [email protected] I will provide some background on the CRISPR/Cas9 technology, some of the rationale for how we came to develop and use this tool, and I will address immediate questions concerning the specificity of the technology. I will also discuss some of the more interest- ing applications. Figure 1 reflects how the cost of DNA sequencing has decreased dramatically over the past two decades due to technological progress. As a result, there has been an explo- sion of data, not only in the sequences of different species, but in sequence differences between individuals within species, between cell types and between diseased and healthy cells. It suffices to say that this is an exciting time to be working in the field of genome engineering. Genome Engineering Typically, genome engineering is achieved by leveraging the cell’s own repair machinery. This can come from the error-prone NHEJ pathway that leads to insertion/deletion (in- del) mutations, which can be used to knock out genes, or, alternatively, we can supply a repair template to overwrite the site of a double-stranded break (DSB) for more-precise genome engineering via the HDR pathway (Figure 2). Figure 1. Advances in DNA-sequencing technologies. (Stratton MR et al., 2009) When we started working on CRISPR/Cas technology1, several well developed tools were already being used—and still are being used—to achieve impressive results in bio- technology, medicine, agriculture, and other fields. At the outset, we were interested in developing an alternative technology to make cloning easier at lower cost with greater scalability. -

Development and Applications of CRISPR-Cas9 for Genome Engineering
Leading Edge Review Development and Applications of CRISPR-Cas9 for Genome Engineering Patrick D. Hsu,1,2,3 Eric S. Lander,1 and Feng Zhang1,2,* 1Broad Institute of MIT and Harvard, 7 Cambridge Center, Cambridge, MA 02141, USA 2McGovern Institute for Brain Research, Department of Brain and Cognitive Sciences, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA 3Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138, USA *Correspondence: [email protected] http://dx.doi.org/10.1016/j.cell.2014.05.010 Recent advances in genome engineering technologies based on the CRISPR-associated RNA- guided endonuclease Cas9 are enabling the systematic interrogation of mammalian genome function. Analogous to the search function in modern word processors, Cas9 can be guided to specific locations within complex genomes by a short RNA search string. Using this system, DNA sequences within the endogenous genome and their functional outputs are now easily edited or modulated in virtually any organism of choice. Cas9-mediated genetic perturbation is simple and scalable, empowering researchers to elucidate the functional organization of the genome at the systems level and establish causal linkages between genetic variations and biological phenotypes. In this Review, we describe the development and applications of Cas9 for a variety of research or translational applications while highlighting challenges as well as future directions. Derived from a remarkable microbial defense system, Cas9 is driving innovative applications from basic biology to biotechnology and medicine. Introduction of the genome and its functions. In biotechnology, precise The development of recombinant DNA technology in the 1970s manipulation of genetic building blocks and regulatory machin- marked the beginning of a new era for biology. -

Recombinant S. Pyogenes CRISPR-Cas9 Catalog Number: 9957-C9
Recombinant S. pyogenes CRISPR-Cas9 Catalog Number: 9957-C9 DESCRIPTION Source E. coli-derived s. pyogenes CRISPR-Cas9 protein S. pyogenes CRISPR-Cas9 KRPAATKKAGQAKK- APKKKRKVGIHGVPAA (Asp2-Asp1368) HHHHHH KKGYGRKKRRQRRRG Accession # Q99ZW2 N-terminus C-terminus N-terminal Sequence Ala Analysis Predicted Molecular 164 kDa Mass SPECIFICATIONS SDS-PAGE 133 kDa, reducing conditions Activity Measured by its ability to cleave a targeted DNA substrate. S. pyogenes CRISPR-Cas9 achieves >80% substrate cleavage, as measured under the described conditions. Endotoxin Level <0.10 EU per 1 μg of the protein by the LAL method. Purity >95%, by SDS-PAGE visualized with Silver Staining and quantitative densitometry by Coomassie® Blue Staining. Formulation Supplied as a 0.2 μm filtered solution in Tris, NaCl, EDTA, Glycerol and TCEP. See Certificate of Analysis for details. Activity Assay Protocol Materials Assay Buffer: 50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 100 µg/mL BSA, pH 7.9 Recombinant Streptococcus pyogenes CRISPR-Cas9 (rS. pyogenes Cas9) (Catalog # 9957-C9) PBR322 vector (NEB, Catalog # N3033S) digested with EcoRI-HF (NEB, Catalog # R3101S)* Dharmacon synthetic sgRNA, targeting sequence: GAGGCAGACAAGGTATAGGG Ethidium Bromide, 10 mg/mL (Amresco, Catalog # X328) Ultrapure DNase/RNase-Free Distilled Water (Invitrogen, Catalog # 10977015), to prepare Assay Buffer DNA gel *Digest was gel purified using gel purification kit and eluted in EB buffer (10 mM Tris-HCl, pH 8.5). Assay 1. Prepare RNP Complex: a. 600 nM sgRNA (6 µL addition from 3 µM stock prepared in Assay Buffer) b. 0.25 μg rS. pyogenes Cas9 c. Add Assay Buffer for a final RNP Complex volume of 26.5 µL d. -

CRISPR/Cas9 System and Gene Editing Tools – on Patent Rights, Recent Disputes and Its Potential Commercial Applicability in Biotechnology and Medicine
ISSN 2003-2382 CRISPR/Cas9 system and gene editing tools – On patent rights, recent disputes and its potential commercial applicability in biotechnology and medicine By Thomas Hedner and Jean Lycke ABSTRACT may also find a future use in “de-extinction” of various animals such as the woolly mammoth The CRISPR/Cas9 discovery has emerged as a and passenger pigeon. powerful technology tool to edit genomes, which The recent discoveries and developments have allows researchers, innovators and life science led to extensive patenting efforts, resulting in some entrepreneurs to alter DNA sequences and modify major patent disputes. The extensive patenting may gene function in a range of species. The simplicity, risk creating a scenario, which could hamper the high efficiency and seemingly broad use of the further development of this technology and ultima- CRISPR/Cas9 system has led to hopes that this tely limit full value creation of this technology for disruptive technology may have the potential to major societal and industrial stakeholders. transform important sectors of biotechnology and medicine. The technology will enable users to make changes in the sequence or expression of virtually 1. INTRODUCTION any gene, cell type or organism. The rapid progress The CRISPR technology, which allows researchers to easily in the development of CRISPR/Cas9-based techno- alter DNA sequences and modify gene function has over logies over the past years has been extraordinary. the past decade emerged a simple and powerful tool for In spite of that, many outstanding questions remain editing genomes1 The CRISPR/Cas9 is a system initially to be addressed, and potentially interesting applica- found in bacteria as a mechanism involved in immune tions as well as potential risks yet need to be explored. -

CRISPR/Cas9 Genome Editing Brochure
mirusbio.com Cas9 Target Sequence Guide RNA GENOME EDITING: CRISPR/CAS9 DELIVERY METHODS GENOME EDITING: CRISPR/CAS9 DELIVERY What is CRISPR/Cas9 Genome Editing? The CRISPR/Cas9 system is a powerful tool for genome editing in mammalian cells that allows researchers to generate genetic variants at lower cost and with higher throughput than alternative methods like zinc finger nuclease (ZFN) or transcription activator-like effector nuclease (TALEN) genome editing. Cas9 PAM Genomic DNA Target Sequence Guide RNA crRNA tracrRNA The CRISPR/Cas9 RNP Complex. The CRISPR associated protein 9 (Cas9) endonuclease (blue) is targeted to DNA by a guide RNA (gRNA), which can be supplied as a two-part system consisting of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) or as a single guide RNA (sgRNA), where the crRNA and tracrRNA are connected by a linker (dotted line). Target recognition is facilitated by the protospacer-adjacent motif (PAM). A double strand break (DSB) occurs 3 bp upstream of the PAM. CRISPR Facilitates Multiple Types of Genome Modification Cleavage of Target DNA By Cas9 Deletion Modication Insertion Multiple Genomic Alterations are Possible Following Cleavage of Target DNA by Cas9. Variable length insertions and/ or deletions (indels) can result near the DNA break due to mistakes in DNA repair by the endogenous non-homologous end joining (NHEJ) pathway. These indels frequently result in disruption of gene function. Alternatively, by supplying a DNA repair template, researchers can leverage the homology-directed repair (HDR) pathway to create defined deletions, insertions or other modifications. 2 TO ORDER | Toll Free 888.530.0801 | Direct 608.441.2852 | www.mirusbio.com Glossary of CRISPR Terms Term Definition CRISPR Associated Protein 9 - Cas9 is an RNA-guided DNA endonuclease from the type Cas9 II CRISPR system of Streptococcus pyogenes that has been adapted for use in genome editing applications. -

Engineering of Primary Human B Cells with CRISPR/Cas9 Targeted Nuclease Received: 26 January 2018 Matthew J
www.nature.com/scientificreports OPEN Engineering of Primary Human B cells with CRISPR/Cas9 Targeted Nuclease Received: 26 January 2018 Matthew J. Johnson1,2,3, Kanut Laoharawee1,2,3, Walker S. Lahr1,2,3, Beau R. Webber1,2,3 & Accepted: 23 July 2018 Branden S. Moriarity1,2,3 Published: xx xx xxxx B cells ofer unique opportunities for gene therapy because of their ability to secrete large amounts of protein in the form of antibody and persist for the life of the organism as plasma cells. Here, we report optimized CRISPR/Cas9 based genome engineering of primary human B cells. Our procedure involves enrichment of CD19+ B cells from PBMCs followed by activation, expansion, and electroporation of CRISPR/Cas9 reagents. We are able expand total B cells in culture 10-fold and outgrow the IgD+ IgM+ CD27− naïve subset from 35% to over 80% of the culture. B cells are receptive to nucleic acid delivery via electroporation 3 days after stimulation, peaking at Day 7 post stimulation. We tested chemically modifed sgRNAs and Alt-R gRNAs targeting CD19 with Cas9 mRNA or Cas9 protein. Using this system, we achieved genetic and protein knockout of CD19 at rates over 70%. Finally, we tested sgRNAs targeting the AAVS1 safe harbor site using Cas9 protein in combination with AAV6 to deliver donor template encoding a splice acceptor-EGFP cassette, which yielded site-specifc integration frequencies up to 25%. The development of methods for genetically engineered B cells opens the door to a myriad of applications in basic research, antibody production, and cellular therapeutics. -

User Method: Alt-R CRISPR-Cas9 RNP Complexes for Microinjection Into B
user method genome editing Microinjection of Bactrocera tryoni (Queensland fruit fly) embryos How to prepare Alt-R® CRISPR-Cas9 ribonucleoprotein complexes for microinjection Contributed by Amanda Choo, University of Adelaide, Adelaide, South Australia The method presented here is provided by customers who have used the Alt-R CRISPR-Cas9 System. This can serve as a starting point for using the Alt-R CRISPR-Cas9 System in similar biological systems, but may not be fully optimized for your gene or application. IDT does not guarantee these methods, and application specialists at IDT can only provide general guidance with limited troubleshooting and support. Materials Kits and reagents Ordering information Enzyme dilution reagents: Option 1: HEPES and KCl General laboratory supplier Option 2: 1X Phosphate buffered saline (PBS) General laboratory supplier Option 3: Opti-MEM® media Thermo Fisher (cat # 51985091) IDT predesigned and custom crRNA: Alt-R CRISPR-Cas9 crRNA www.idtdna.com/CRISPR-Cas9 Alt-R CRISPR-Cas9 tracrRNA IDT (cat # 1072532, 1072533, 1072534) or Alt-R CRISPR-Cas9 tracrRNA – ATTO™ 550 IDT (cat # 1075927, 1075928) Alt-R S.p. Cas9 Nuclease 3NLS IDT (cat # 1074181, 1074182) Nuclease-Free Duplex Buffer IDT (cat # 11-01-03-01) Reagents for 10X Injection buffer: Sodium phosphate General laboratory supplier KCl General laboratory supplier Nuclease-Free Water IDT (cat # 11-05-01-14) HDR template: Ultramer® DNA Oligonucleotides IDT (www.idtdna.com/Ultramer) See what more we can do for you at www.idtdna.com. user method genome editing Methods A. Prepare buffers and enzyme 1. Prepare the Cas9 enzyme working buffer as described in the following table: Component Amount Final concentration 1 M HEPES, pH 7.5 200 µL 20 mM 1 M KCl 1.5 mL 150 mM Nuclease-Free Water ~6.5 mL* — Final volume 10 mL — * Add 6.5 mL of water, verify pH 7.5, and add more water to reach final volume. -

CRISPR-Cas9 DNA Base-Editing and Prime-Editing
International Journal of Molecular Sciences Review CRISPR-Cas9 DNA Base-Editing and Prime-Editing Ariel Kantor 1,2,*, Michelle E. McClements 1,2 and Robert E. MacLaren 1,2 1 Nuffield Laboratory of Ophthalmology, Nuffield Department of Clinical Neurosciences & NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford OX3 9DU, UK 2 Oxford Eye Hospital, Oxford University Hospitals NHS Foundation Trust, Oxford OX3 9DU, UK * Correspondence: [email protected] Received: 14 July 2020; Accepted: 25 August 2020; Published: 28 August 2020 Abstract: Many genetic diseases and undesirable traits are due to base-pair alterations in genomic DNA. Base-editing, the newest evolution of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas-based technologies, can directly install point-mutations in cellular DNA without inducing a double-strand DNA break (DSB). Two classes of DNA base-editors have been described thus far, cytosine base-editors (CBEs) and adenine base-editors (ABEs). Recently, prime-editing (PE) has further expanded the CRISPR-base-edit toolkit to all twelve possible transition and transversion mutations, as well as small insertion or deletion mutations. Safe and efficient delivery of editing systems to target cells is one of the most paramount and challenging components for the therapeutic success of BEs. Due to its broad tropism, well-studied serotypes, and reduced immunogenicity, adeno-associated vector (AAV) has emerged as the leading platform for viral delivery of genome editing agents, including DNA-base-editors. In this review, we describe the development of various base-editors, assess their technical advantages and limitations, and discuss their therapeutic potential to treat debilitating human diseases. -

CRISPR-Cas9 Gene Editing and Rapid Detection of Gene-Edited Mutants Using High-Resolution
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.04.428760; this version posted February 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 CRISPR-Cas9 gene editing and rapid detection of gene-edited mutants using high-resolution 2 melting in the apple scab fungus, Venturia inaequalis 3 4 Mercedes Rocafort1, Saadiah Arshed2, Debbie Hudson3, Jaspreet Singh3, Joanna K. Bowen2, 5 Kim M. Plummer4, Rosie E. Bradshaw5,6, Richard D. Johnson3, Linda J. Johnson3 and Carl H. 6 Mesarich1,6,* 7 8 1Laboratory of Molecular Plant Pathology, School of Agriculture and Environment, Massey 9 University, Palmerston North 4410, New Zealand. 10 2The New Zealand Institute for Plant and Food Research Limited, Mount Albert Research 11 Centre, Auckland 1025, New Zealand. 12 3Grasslands Research Centre, AgResearch Limited, Palmerston North 4410, New Zealand. 13 4Department of Animal, Plant and Soil Sciences, La Trobe University, AgriBio, Centre for 14 AgriBiosciences, La Trobe University, Bundoora, Victoria 3086, Australia. 15 5Laboratory of Molecular Plant Pathology, School of Fundamental Sciences, Massey 16 University, Palmerston North 4410, New Zealand. 17 6The New Zealand Bio-Protection Research Centre, Massey University, Palmerston North 18 4410, New Zealand. 19 20 *Corresponding author: Carl H. Mesarich: [email protected] 21 22 23 24 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.02.04.428760; this version posted February 5, 2021. -

What Is CRISPR/Cas9? Page 5
Your Guide to Understanding CRISPR 1 Contact [email protected] | (888) 611-6883 20161130 TABLE OF CONTENTS Introduction Page 3 A Brief History of CRISPR Page 3 Gene Editing Before CRISPR Page 4 What is CRISPR/Cas9? Page 5 CRISPR Applications Page 7 CRISPR Guides Page 9 Developments in Synthetic sgRNA Page 12 Alternatives to S. pyogenes Cas9 Page 13 Gene Editing is Just the Beginning Page 14 2 Contact [email protected] | (888) 611-6883 20161130 INTRODUCTION CRISPR is igniting a revolution. A relatively recent discovery in the timeline of biotechnology, CRISPR is quickly becoming a standard and flexible laboratory tool, and it is well on its way to permeating a large variety of applications. Researchers are deploying CRISPR across a wide range of life science disciplines, from agriculture and medicine to biofuels and industrial fermentation. Read on for a crash course in everything you need to know if you’re just getting your first taste of CRISPR. A BRIEF HISTORY OF CRISPR The foundational discoveries that led to CRISPR/Cas9 technology can be traced back to 1993, when the genomic regions known as CRISPR loci were first identified. In 2007, after years of studying CRISPR genetic motifs, researchers came to the conclusion that CRISPR’s function is related to microbial cellular immunity. Throughout the next 5 years, several research groups worked to elucidate the underlying molecular mechanisms behind CRISPR in Prokaryotes. CRISPR works as a form of Prokaryotic immunity that identifies, targets, and eliminates bacteriophage and foreign DNA. By 2012, researchers realized that CRISPR could be adapted for engineering the genomes of microbes, plants, animal, and other varieties of cells. -
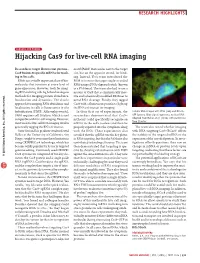
Sensors and Probes: Hijacking Cas9 for Live-Cell RNA Imaging
RESEARCH HIGHLIGHTS SENSORS AND PROBES Hijacking Cas9 for live-cell RNA imaging Researchers target fluorescent protein– motif (PAM) that resides next to the target Cas9 fusions to specific mRNAs for track- site, but on the opposite strand, for bind- ing in live cells. ing. Instead, Yeo’s team introduced the RNAs are a vitally important class of bio- PAM in trans to the target single-stranded molecules that function at every level of RNA using a DNA oligonucleotide (known gene expression. However, tools for imag- as a PAMmer). The team also had to use a ing RNA in living cells lag behind analogous mutant of Cas9 that is enzymatically inac- methods for imaging protein abundance, tive and a chemically modified PAMmer to localization and dynamics. The classic avoid RNA cleavage. Finally, they tagged approach for imaging RNA abundance and Cas9 with a fluorescent protein to light up localization in cells is fluorescence in situ the RNAs of interest for imaging. hybridization (FISH). Although powerful, In their first set of experiments, the Cellular RNA imaged with FISH (red) and RCas9– FISH requires cell fixation, which is not researchers demonstrated that Cas9– GFP (green). Blue signal represents nuclear DNA. Adapted from Nelles et al. (2016) with permission compatible with live-cell imaging. However, mCherry could specifically recognize an from Elsevier. most tools for live-cell RNA imaging involve mRNA in the cell’s nucleus and then be genetically tagging the RNA of interest. properly exported into the cytoplasm along The team also tested whether imaging Gene Yeo and his graduate student David with the RNA.