Phylogenetic and Biochemical Evidence for Sterol Synthesis in the Bacterium Gemmata Obscuriglobus
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Investigation of Histone Lysine-Specific Demethylase 5D
FULL LENGTH Iranian Biomedical Journal 20(2): 117-121 April 2016 Investigation of Histone Lysine-Specific Demethylase 5D (KDM5D) Isoform Expression in Prostate Cancer Cell Lines: a System Approach Zohreh Jangravi1, 2, Mohammad Najafi1,3 and Mohammd Shabani*1 1Dept. of Biochemistry, Iran University of Medical Sciences, Tehran, Iran; 2Dept. of Molecular Systems Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran; 3Dept. of Biochemistry, Razi Drug Research Center, Iran University of Medical Sciences, Tehran, Iran Received 2 September 2014; revised 3 December 2014; accepted 7 December 2014 ABSTRACT Background: It is now well-demonstrated that histone demethylases play an important role in developmental controls, cell-fate decisions, and a variety of diseases such as cancer. Lysine-specific demethylase 5D (KDM5D) is a male-specific histone demethylase that specifically demethylates di- and tri-methyl H3K4 at the start site of active gene. In this light, the aim of this study was to investigate isoform/transcript-specific expression profiles of KDM5D in three prostate cancer cell lines, Du-145, LNCaP, and PC3. Methods: Real-time PCR analysis was performed to determine the expression levels of different KDM5D transcripts in the prostate cell lines. A gene regulatory network was established to analyze the gene expression profile. Results: Significantly different expression levels of both isoforms were found among the three cell lines. Interestingly, isoform I was expressed in three cell lines while isoform III did only in DU-145. The expression levels of both isoforms were higher in DU-145 when compared to other cell lines (P<0.0001). -

Lanosterol 14Α-Demethylase (CYP51)
463 Lanosterol 14-demethylase (CYP51), NADPH–cytochrome P450 reductase and squalene synthase in spermatogenesis: late spermatids of the rat express proteins needed to synthesize follicular fluid meiosis activating sterol G Majdicˇ, M Parvinen1, A Bellamine2, H J Harwood Jr3, WWKu3, M R Waterman2 and D Rozman4 Veterinary Faculty, Clinic of Reproduction, Cesta v Mestni log 47a, 1000 Ljubljana, Slovenia 1Institute of Biomedicine, Department of Anatomy, University of Turku, Kiinamyllynkatu 10, FIN-20520 Turku, Finland 2Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, Tennessee 37232–0146, USA 3Pfizer Central Research, Department of Metabolic Diseases, Box No. 0438, Eastern Point Road, Groton, Connecticut 06340, USA 4Institute of Biochemistry, Medical Center for Molecular Biology, Medical Faculty University of Ljubljana, Vrazov trg 2, SI-1000 Ljubljana, Slovenia (Requests for offprints should be addressed to D Rozman; Email: [email protected]) (G Majdicˇ is now at Department of Internal Medicine, UT Southwestern Medical Center, Dallas, Texas 75235–8857, USA) Abstract Lanosterol 14-demethylase (CYP51) is a cytochrome detected in step 3–19 spermatids, with large amounts in P450 enzyme involved primarily in cholesterol biosynthe- the cytoplasm/residual bodies of step 19 spermatids, where sis. CYP51 in the presence of NADPH–cytochrome P450 P450 reductase was also observed. Squalene synthase was reductase converts lanosterol to follicular fluid meiosis immunodetected in step 2–15 spermatids of the rat, activating sterol (FF-MAS), an intermediate of cholesterol indicating that squalene synthase and CYP51 proteins are biosynthesis which accumulates in gonads and has an not equally expressed in same stages of spermatogenesis. additional function as oocyte meiosis-activating substance. -

Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae
Journal of Fungi Article Characterization of the Ergosterol Biosynthesis Pathway in Ceratocystidaceae Mohammad Sayari 1,2,*, Magrieta A. van der Nest 1,3, Emma T. Steenkamp 1, Saleh Rahimlou 4 , Almuth Hammerbacher 1 and Brenda D. Wingfield 1 1 Department of Biochemistry, Genetics and Microbiology, Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria 0002, South Africa; [email protected] (M.A.v.d.N.); [email protected] (E.T.S.); [email protected] (A.H.); brenda.wingfi[email protected] (B.D.W.) 2 Department of Plant Science, University of Manitoba, 222 Agriculture Building, Winnipeg, MB R3T 2N2, Canada 3 Biotechnology Platform, Agricultural Research Council (ARC), Onderstepoort Campus, Pretoria 0110, South Africa 4 Department of Mycology and Microbiology, University of Tartu, 14A Ravila, 50411 Tartu, Estonia; [email protected] * Correspondence: [email protected]; Fax: +1-204-474-7528 Abstract: Terpenes represent the biggest group of natural compounds on earth. This large class of organic hydrocarbons is distributed among all cellular organisms, including fungi. The different classes of terpenes produced by fungi are mono, sesqui, di- and triterpenes, although triterpene ergosterol is the main sterol identified in cell membranes of these organisms. The availability of genomic data from members in the Ceratocystidaceae enabled the detection and characterization of the genes encoding the enzymes in the mevalonate and ergosterol biosynthetic pathways. Using Citation: Sayari, M.; van der Nest, a bioinformatics approach, fungal orthologs of sterol biosynthesis genes in nine different species M.A.; Steenkamp, E.T.; Rahimlou, S.; of the Ceratocystidaceae were identified. -

Sterol Homeostasis Requires Regulated Degradation of Squalene
RESEARCH ARTICLE elife.elifesciences.org Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4 Ombretta Foresti1,2, Annamaria Ruggiano1,2, Hans K Hannibal-Bach3, Christer S Ejsing3, Pedro Carvalho1,2* 1Cell and Developmental Biology Programme, Center for Genomic Regulation (CRG), Barcelona, Spain; 2Universitat Pompeu Fabra, Barcelona, Spain; 3Department of Biochemistry and Molecular Biology, University of Southern Denmark, Odense, Denmark Abstract Sterol homeostasis is essential for the function of cellular membranes and requires feedback inhibition of HMGR, a rate-limiting enzyme of the mevalonate pathway. As HMGR acts at the beginning of the pathway, its regulation affects the synthesis of sterols and of other essential mevalonate-derived metabolites, such as ubiquinone or dolichol. Here, we describe a novel, evolutionarily conserved feedback system operating at a sterol-specific step of the mevalonate pathway. This involves the sterol-dependent degradation of squalene monooxygenase mediated by the yeast Doa10 or mammalian Teb4, a ubiquitin ligase implicated in a branch of the endoplasmic reticulum (ER)-associated protein degradation (ERAD) pathway. Since the other branch of ERAD is required for HMGR regulation, our results reveal a fundamental role for ERAD in sterol homeostasis, with the two branches of this pathway acting together to control sterol biosynthesis at different levels and thereby allowing independent regulation of multiple products of the mevalonate pathway. DOI: 10.7554/eLife.00953.001 *For correspondence: pedro. [email protected] Introduction Sterols, such as cholesterol in animals or ergosterol in yeast, are essential components of cellular Competing interests: The authors membranes and their concentration to a large extent determines many of the membrane properties, declare that no competing interests exist. -

X Chromosome Dosage of Histone Demethylase KDM5C Determines Sex Differences in Adiposity
X chromosome dosage of histone demethylase KDM5C determines sex differences in adiposity Jenny C. Link, … , Arthur P. Arnold, Karen Reue J Clin Invest. 2020. https://doi.org/10.1172/JCI140223. Research Article Genetics Metabolism Graphical abstract Find the latest version: https://jci.me/140223/pdf The Journal of Clinical Investigation RESEARCH ARTICLE X chromosome dosage of histone demethylase KDM5C determines sex differences in adiposity Jenny C. Link,1 Carrie B. Wiese,2 Xuqi Chen,3 Rozeta Avetisyan,2 Emilio Ronquillo,2 Feiyang Ma,4 Xiuqing Guo,5 Jie Yao,5 Matthew Allison,6 Yii-Der Ida Chen,5 Jerome I. Rotter,5 Julia S. El -Sayed Moustafa,7 Kerrin S. Small,7 Shigeki Iwase,8 Matteo Pellegrini,4 Laurent Vergnes,2 Arthur P. Arnold,3 and Karen Reue1,2 1Molecular Biology Institute, 2Human Genetics, David Geffen School of Medicine, 3Integrative Biology and Physiology, and 4Molecular, Cellular and Developmental Biology, UCLA, Los Angeles, California, USA. 5Institute for Translational Genomics and Population Sciences, Department of Pediatrics, Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, California, USA. 6Division of Preventive Medicine, School of Medicine, UCSD, San Diego, California, USA. 7Department of Twin Research and Genetic Epidemiology, King’s College London, London, United Kingdom. 8Human Genetics, Medical School, University of Michigan, Ann Arbor, Michigan, USA. Males and females differ in body composition and fat distribution. Using a mouse model that segregates gonadal sex (ovaries and testes) from chromosomal sex (XX and XY), we showed that XX chromosome complement in combination with a high-fat diet led to enhanced weight gain in the presence of male or female gonads. -

Targeting JARID1B's Demethylase Activity Blocks a Subset of Its Functions in Oral Cancer
www.impactjournals.com/oncotarget/ Oncotarget, 2018, Vol. 9, (No. 10), pp: 8985-8998 Research Paper Targeting JARID1B's demethylase activity blocks a subset of its functions in oral cancer Nicole D. Facompre1, Kayla M. Harmeyer1, Varun Sahu1, Phyllis A. Gimotty2, Anil K. Rustgi3, Hiroshi Nakagawa3 and Devraj Basu1,4,5 1Department of Otorhinolaryngology, Head and Neck Surgery, The University of Pennsylvania, Philadelphia, PA, USA 2Department of Biostatistics Epidemiology and Informatics, The University of Pennsylvania, Philadelphia, PA, USA 3Department of Medicine, The University of Pennsylvania, Philadelphia, PA, USA 4Philadelphia VA Medical Center, Philadelphia, PA, USA 5The Wistar Institute, Philadelphia, PA, USA Correspondence to: Devraj Basu, email: [email protected] Keywords: oral cancer; JARID1B; cancer stem cells; E-cadherin Received: July 04, 2017 Accepted: October 13, 2017 Published: December 15, 2017 Copyright: Facompre et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Upregulation of the H3K4me3 demethylase JARID1B is linked to acquisition of aggressive, stem cell-like features by many cancer types. However, the utility of emerging JARID1 family inhibitors remains uncertain, in part because JARID1B's functions in normal development and malignancy are diverse and highly context- specific. In this study, responses of oral squamous cell carcinomas (OSCCs) to catalytic inhibition of JARID1B were assessed using CPI-455, the first tool compound with true JARID1 family selectivity. CPI-455 attenuated clonal sphere and tumor formation by stem-like cells that highly express JARID1B while also depleting the CD44-positive and Aldefluor-high fractions conventionally used to designate OSCC stem cells. -

Biodegradation of Benzidine Based Dye Direct Blue-6 by Pseudomonas Desmolyticum NCIM 2112
ARTICLE IN PRESS Bioresource Technology xxx (2006) xxx–xxx Biodegradation of benzidine based dye Direct Blue-6 by Pseudomonas desmolyticum NCIM 2112 S.D. Kalme, G.K. Parshetti, S.U. Jadhav, S.P. Govindwar * Department of Biochemistry, Shivaji University, Kolhapur 416004, India Received 16 March 2006; received in revised form 12 May 2006; accepted 16 May 2006 Abstract Pseudomonas desmolyticum NCIM 2112 was able to degrade a diazo dye Direct Blue-6 (100 mg lÀ1) completely within 72 h of incubation with 88.95% reduction in COD in static anoxic condition. Induction in the activity of oxidative enzymes (LiP, laccase) and tyrosinase while decolorization in the batch culture represents their role in degradation. Dye also induced the activity of aminopyrine N-demethylase, one of the enzyme of mixed function oxidase system. The biodegradation was monitored by UV–Vis, IR spectroscopy and HPLC. The final products, 4-amino naphthalene and amino naphthalene sulfonic acid were characterized by GC–mass spectroscopy. Ó 2006 Elsevier Ltd. All rights reserved. Keywords: Direct Blue-6; Pseudomonas desmolyticum; Biodegradation; Azo dye; Lignin peroxidase 1. Introduction of sulfur containing dyes by using white rot fungus Dichomitus squalens and Coriolus versicolor (Eichlerova´ Azo dyes account for the majority of all textile dyestuffs et al., 2006; Sanghi et al., 2006). The decolorization of sul- produced and are the most commonly used synthetic dyes fonated azo dyes by different pseudomonas species without in the textile, food, paper making, color paper printing, shaking condition (anaerobic incubation of azo dyes) has leather and cosmetic industries (Chang et al., 2001). Sulfo- been reported earlier (Banat et al., 1996; Puvaneshwari nated azo dyes, the largest class of dyes, have great struc- et al., 2002). -

Aquatic Toxicology 176 (2016) 116–127
Aquatic Toxicology 176 (2016) 116–127 Contents lists available at ScienceDirect Aquatic Toxicology j ournal homepage: www.elsevier.com/locate/aquatox Linking the response of endocrine regulated genes to adverse effects on sex differentiation improves comprehension of aromatase inhibition in a Fish Sexual Development Test a,∗ b b b Elke Muth-Köhne , Kathi Westphal-Settele , Jasmin Brückner , Sabine Konradi , c a a,1 d,1 Viktoria Schiller , Christoph Schäfers , Matthias Teigeler , Martina Fenske a Fraunhofer IME, Department of Ecotoxicology, Auf dem Aberg 1, 57392 Schmallenberg, Germany b German Environment Agency (UBA), Woerlitzer Platz 1, 06844 Dessau, Germany c Fraunhofer IME, Attract Group UNIFISH, Forckenbeckstraße 6, 52074 Aachen, Germany d Fraunhofer IME, Project Group Translational Medicine and Pharmacology TMP, Forckenbeckstraße 6, 52074 Aachen, Germany a r a t i c l e i n f o b s t r a c t Article history: The Fish Sexual Development Test (FSDT) is a non-reproductive test to assess adverse effects of endocrine Received 23 December 2015 disrupting chemicals. With the present study it was intended to evaluate whether gene expression end- Received in revised form 13 April 2016 points would serve as predictive markers of endocrine disruption in a FSDT. For proof-of-concept, a FSDT Accepted 19 April 2016 according to the OECD TG 234 was conducted with the non-steroidal aromatase inhibitor fadrozole (test Available online 20 April 2016 concentrations: 10 g/L, 32 g/L, 100 g/L) using zebrafish (Danio rerio). Gene expression analyses using quantitative RT-PCR were included at 48 h, 96 h, 28 days and 63 days post fertilization (hpf, dpf). -

Evolution of the Tyrosinase Gene Family in Bivalve Molluscs: Independent Ex- Pansion of the Mantle Gene Repertoire
Accepted Manuscript Evolution of the tyrosinase gene family in bivalve molluscs: independent ex- pansion of the mantle gene repertoire Felipe Aguilera, Carmel McDougall, Bernard M Degnan PII: S1742-7061(14)00151-2 DOI: http://dx.doi.org/10.1016/j.actbio.2014.03.031 Reference: ACTBIO 3182 To appear in: Acta Biomaterialia Please cite this article as: Aguilera, F., McDougall, C., Degnan, B.M., Evolution of the tyrosinase gene family in bivalve molluscs: independent expansion of the mantle gene repertoire, Acta Biomaterialia (2014), doi: http:// dx.doi.org/10.1016/j.actbio.2014.03.031 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. Evolution of the tyrosinase gene family in bivalve molluscs: independent expansion of the mantle gene repertoire Authors: Felipe Aguilera, Carmel McDougall and Bernard M Degnan* Authors Affiliations: Centre for Marine Sciences, School of Biological Sciences, The University of Queensland, Brisbane, Australia, 4072. Corresponding author: Bernard M Degnan, Centre for Marine Sciences, School of Biological Sciences, The University of Queensland, Brisbane, Australia, 4072. Ph: (+61) 7 3365 2467 Fax: (+61+ 7 3365 1199. Email. [email protected] Page 1 Abstract Tyrosinase is a copper-containing enzyme that mediates the hydroxylation of monophenols and oxidation of o-diphenols to o-quinones. -

Steroidal Triterpenes of Cholesterol Synthesis
Molecules 2013, 18, 4002-4017; doi:10.3390/molecules18044002 OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules Review Steroidal Triterpenes of Cholesterol Synthesis Jure Ačimovič and Damjana Rozman * Centre for Functional Genomics and Bio-Chips, Faculty of Medicine, Institute of Biochemistry, University of Ljubljana, Zaloška 4, Ljubljana SI-1000, Slovenia; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +386-1-543-7591; Fax: +386-1-543-7588. Received: 18 February 2013; in revised form: 19 March 2013 / Accepted: 27 March 2013 / Published: 4 April 2013 Abstract: Cholesterol synthesis is a ubiquitous and housekeeping metabolic pathway that leads to cholesterol, an essential structural component of mammalian cell membranes, required for proper membrane permeability and fluidity. The last part of the pathway involves steroidal triterpenes with cholestane ring structures. It starts by conversion of acyclic squalene into lanosterol, the first sterol intermediate of the pathway, followed by production of 20 structurally very similar steroidal triterpene molecules in over 11 complex enzyme reactions. Due to the structural similarities of sterol intermediates and the broad substrate specificity of the enzymes involved (especially sterol-Δ24-reductase; DHCR24) the exact sequence of the reactions between lanosterol and cholesterol remains undefined. This article reviews all hitherto known structures of post-squalene steroidal triterpenes of cholesterol synthesis, their biological roles and the enzymes responsible for their synthesis. Furthermore, it summarises kinetic parameters of enzymes (Vmax and Km) and sterol intermediate concentrations from various tissues. Due to the complexity of the post-squalene cholesterol synthesis pathway, future studies will require a comprehensive meta-analysis of the pathway to elucidate the exact reaction sequence in different tissues, physiological or disease conditions. -

Expression of Selected Genes Involved in Steroidogenesis in the Course of Enucleation-Induced Rat Adrenal Regeneration
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 33: 613-623, 2014 Expression of selected genes involved in steroidogenesis in the course of enucleation-induced rat adrenal regeneration MARIANNA TYCZEWSKA, MARCIN RUCINSKI, AGNIESZKA ZIOLKOWSKA, MARCIN TREJTER, MARTA SZYSZKA and LUDWIK K. MALENDOWICZ Department of Histology and Embryology, Poznan University of Medical Sciences, Poznan, Poland Received October 28, 2013; Accepted December 6, 2013 DOI: 10.3892/ijmm.2013.1599 Abstract. The enucleation-induced (EI) rapid proliferation however, throughout the entire experimental period, there were of adrenocortical cells is followed by their differentiation, no statistically significant differences observed. After the initial the degree of which may be characterized by the expression decrease in steroidogenic factor 1 (Sf-1) mRNA levels observed of genes directly and indirectly involved in steroid hormone on the 1st day of the experiment, a marked upregulation in its biosynthesis. In this study, out of 30,000 transcripts of genes expression was observed from there on. Data from the current identified by means of Affymetrix Rat Gene 1.1 ST Array, we study strongly suggest the role of Fabp6, Lipe and Soat1 in aimed to select genes (either up- or downregulated) involved supplying substrates of regenerating adrenocortical cells for in steroidogenesis in the course of enucleation-induced adrenal steroid synthesis. Our results indicate that during the first days regeneration. On day 1, we found 32 genes with altered of adrenal regeneration, intense synthesis of cholesterol may expression levels, 15 were upregulated and 17 were down- occur, which is then followed by its conversion into cholesteryl regulated [i.e., 3β-hydroxysteroid dehydrogenase (Hsd3β), esters. -
Figures Intermediary Ontogeny.Pptx
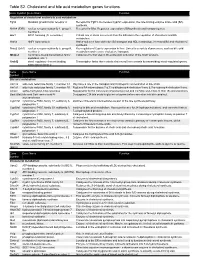
Table S2. Cholesterol and bile acid metabolism genes functions. Gene Symbol Gene Name Function Regulation of cholesterol and/or bile acid metabolism Fgfr4 fibroblast growth factor receptor 4 Receptor for Fgf15. Decreases Cyp7a1 expression, the rate-limiting enzyme in bile acid (BA) synthesis. Nr1h4 (FXR) nuclear receptor subfamily 1, group H, Receptor for BAs. Regulates expression of BAsynthesis and transport genes. member 4 Arv1 ARV1 homolog (S. cerevisiae) Critical role in sterol movement from the ER and in the regulation of cholesterol and BA metabolism. Hnf1a HNF1 homeobox A Hnf1a-null mice have defective BA transport and HDL metabolism, increased BA and cholesterol synthesis. Nr5a2 (Lrh1) nuclear receptor subfamily 5, group A, Key regulator of Cyp7a expression in liver. Linked to a variety of processes, such as bile acid member 2 metabolism and reverse cholesterol transport. Mbtps1 membrane-bound transcription factor Catalyzes the first step in the proteolytic activation of the Srebf proteins. peptidase, site 1 Srebf2 sterol regulatory element binding Transcription factor that controls cholesterol homeostasis by transcribing sterol-regulated genes. transcription factor 2 Gene Gene Name Function Symbol Bile acid metabolism Akr1c6 aldo-keto reductase family 1, member C1 May have a role in the transport and intrahepatic concentration of bile acids. Akr1d1 aldo-keto reductase family 1, member D1 Reduces BA intermediates 7-α,12-α-dihydroxy-4-cholesten-3-one & 7-α-hydroxy-4-cholesten-3-one. Amacr alpha-methylacyl-CoA racemase Responsible for the conversion of pristanoyl-CoA and C27-bile acyl-CoAs to their (S)-stereoisomers. Baat (Bat) bile acid CoA: amino acid N- Conjugates C24 bile acids to glycine or taurine before excretion into bile canaliculi.