Allobacillus Salarius Sp. Nov., and Allobacillus Saliphilus Sp. Nov., Isolated from Shrimp Paste (Ka- Pi) in Thailand
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Lentibacillus Salicampi Gen. Nov., Sp. Nov., a Moderately Halophilic
International Journal of Systematic and Evolutionary Microbiology (2002), 52, 2043–2048 DOI: 10.1099/ijs.0.02335-0 Lentibacillus salicampi gen. nov., sp. nov., a NOTE moderately halophilic bacterium isolated from a salt field in Korea 1 Microbial Genomics Jung-Hoon Yoon,1 Kook Hee Kang2 and Yong-Ha Park1,3 Laboratory, Korea Research Institute of Bioscience and Author for correspondence: Biotechnology (KRIBB), Yong-Ha Park. Tel: j82 42 860 4620. Fax: j82 42 862 1315. PO Box 115, Yusong, e-mail: yhpark!mail.kribb.re.kr Taejon, Korea 2 Department of Food and A Gram-variable, aerobic, endospore-forming, rod-shaped bacterial strain, SF- Life Science, 20T, which was isolated from a salt field in Korea, was subjected to a Sungkyunkwan University, Chunchun-dong 300, polyphasic taxonomic study. Cells of this organism were motile by means of Jangan-gu, Suwon, Korea single flagella. Strain SF-20T grew optimally in the presence of 4–8% NaCl. The 3 National Research cell wall peptidoglycan contained meso-diaminopimelic acid as the diagnostic Laboratory of Molecular diamino acid. The predominant menaquinone is MK-7. Strain SF-20T has a Ecosystematics, Institute of cellular fatty acid profile containing major amounts of branched fatty acids. Probionics, Probionic Corporation, Bio-venture The major fatty acids are anteiso-C15:0 and iso-C16:0. The cellular phospholipids Centre, KRIBB, Yusong, are phosphatidylglycerol and diphosphatidylglycerol. The GMC content of the Taejon, Korea DNA is 44 mol%. Strain SF-20T is phylogenetically closely related to the genus Bacillus and some related genera and, particularly, formed a coherent cluster with the genera Salibacillus and Virgibacillus. -

Genome Sequence of the Moderately Halophilic Yellow Sea Bacterium Lentibacillus Salicampi ATCC BAA-719T
University of New Hampshire University of New Hampshire Scholars' Repository Faculty Publications 7-18-2019 Genome Sequence of the Moderately Halophilic Yellow Sea Bacterium Lentibacillus salicampi ATCC BAA-719T Milto Simoes University of New Hampshire, Manchester Kyle S. MacLea University of New Hampshire, Manchester, [email protected] Follow this and additional works at: https://scholars.unh.edu/faculty_pubs Recommended Citation Simoes, Milto and MacLea, Kyle S., "Genome Sequence of the Moderately Halophilic Yellow Sea Bacterium Lentibacillus salicampi ATCC BAA-719T" (2019). Microbiology Resource Announcements. 702. https://scholars.unh.edu/faculty_pubs/702 This Article is brought to you for free and open access by University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Faculty Publications by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. GENOME SEQUENCES crossm Downloaded from Genome Sequence of the Moderately Halophilic Yellow Sea Bacterium Lentibacillus salicampi ATCC BAA-719T Milto Simoes Junior,a Kyle S. MacLeaa,b,c http://mra.asm.org/ aBiotechnology Program, University of New Hampshire, Manchester, New Hampshire, USA bBiology Program, University of New Hampshire, Manchester, New Hampshire, USA cDepartment of Life Sciences, University of New Hampshire, Manchester, New Hampshire, USA ABSTRACT Lentibacillus salicampi SF-20T (ϭATCC BAA-719T) was first isolated from a Yellow Sea salt field in Korea in 2002. Here, we report that the L. salicampi ATCC BAA-719T genome sequence has a predicted length of 3,897,716 bp, containing on November 15, 2019 at UNIVERSITY OF NEW HAMPSHIRE LIBRARY 3,945 total genes and a CRISPR array, with a GϩC content of 43.0%. -

Assessment of Chicken Carcass Microbiome Responses During
www.nature.com/scientificreports OPEN Assessment of Chicken Carcass Microbiome Responses During Processing in the Presence of Received: 26 September 2016 Accepted: 23 January 2017 Commercial Antimicrobials Using Published: 23 February 2017 a Next Generation Sequencing Approach Sun Ae Kim1,*, Si Hong Park1,*, Sang In Lee1, Casey M. Owens2 & Steven C. Ricke1 The purpose of this study was to 1) identify microbial compositional changes on chicken carcasses during processing, 2) determine the antimicrobial efficacy of peracetic acid (PAA) and Amplon (blend of sulfuric acid and sodium sulfate) at a poultry processing pilot plant scale, and 3) compare microbial communities between chicken carcass rinsates and recovered bacteria from media. Birds were collected from each processing step and rinsates were applied to estimate aerobic plate count (APC) and Campylobacter as well as Salmonella prevalence. Microbiome sequencing was utilized to identify microbial population changes over processing and antimicrobial treatments. Only the PAA treatment exhibited significant reduction of APC at the post chilling step while both Amplon and PAA yielded detectable Campylobacter reductions at all steps. Based on microbiome sequencing, Firmicutes were the predominant bacterial group at the phyla level with over 50% frequency in all steps while the relative abundance of Proteobacteria decreased as processing progressed. Overall microbiota between rinsate and APC plate microbial populations revealed generally similar patterns at the phyla level but they were different at the genus level. Both antimicrobials appeared to be effective on reducing problematic bacteria and microbiome can be utilized to identify optimal indicator microorganisms for enhancing product quality. Chickens and other poultry products are some of the most popular primary food products throughout the world1. -

Isolation and Diversity of Sediment Bacteria in The
bioRxiv preprint doi: https://doi.org/10.1101/638304; this version posted May 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Isolation and Diversity of Sediment Bacteria in the 2 Hypersaline Aiding Lake, China 3 4 Tong-Wei Guan, Yi-Jin Lin, Meng-Ying Ou, Ke-Bao Chen 5 6 7 Institute of Microbiology, Xihua University, Chengdu 610039, P. R. China. 8 9 Author for correspondence: 10 Tong-Wei Guan 11 Tel/Fax: +86 028 87720552 12 E-mail: [email protected] 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 bioRxiv preprint doi: https://doi.org/10.1101/638304; this version posted May 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 29 Abstract A total of 343 bacteria from sediment samples of Aiding Lake, China, were isolated using 30 nine different media with 5% or 15% (w/v) NaCl. The number of species and genera of bacteria recovered 31 from the different media significantly varied, indicating the need to optimize the isolation conditions. 32 The results showed an unexpected level of bacterial diversity, with four phyla (Firmicutes, 33 Actinobacteria, Proteobacteria, and Rhodothermaeota), fourteen orders (Actinopolysporales, 34 Alteromonadales, Bacillales, Balneolales, Chromatiales, Glycomycetales, Jiangellales, Micrococcales, 35 Micromonosporales, Oceanospirillales, Pseudonocardiales, Rhizobiales, Streptomycetales, and 36 Streptosporangiales), including 17 families, 41 genera, and 71 species. -

Insights Into Halophilic Microbial Adaptation: Analysis of Integrons and Associated Genomic Structures and Characterization of A
American University in Cairo AUC Knowledge Fountain Theses and Dissertations Student Research Summer 8-25-2021 Insights Into Halophilic Microbial Adaptation: Analysis of Integrons and Associated Genomic Structures and Characterization of a Nitrilase in Hypersaline Environments Sarah Sonbol [email protected] Follow this and additional works at: https://fount.aucegypt.edu/etds Part of the Bacteriology Commons, Biochemistry Commons, Biodiversity Commons, Bioinformatics Commons, Biotechnology Commons, Environmental Microbiology and Microbial Ecology Commons, Genomics Commons, Marine Biology Commons, Molecular Biology Commons, and the Molecular Genetics Commons Recommended Citation APA Citation Sonbol, S. (2021).Insights Into Halophilic Microbial Adaptation: Analysis of Integrons and Associated Genomic Structures and Characterization of a Nitrilase in Hypersaline Environments [Doctoral Dissertation, the American University in Cairo]. AUC Knowledge Fountain. https://fount.aucegypt.edu/etds/1673 MLA Citation Sonbol, Sarah. Insights Into Halophilic Microbial Adaptation: Analysis of Integrons and Associated Genomic Structures and Characterization of a Nitrilase in Hypersaline Environments. 2021. American University in Cairo, Doctoral Dissertation. AUC Knowledge Fountain. https://fount.aucegypt.edu/etds/1673 This Doctoral Dissertation is brought to you for free and open access by the Student Research at AUC Knowledge Fountain. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of AUC Knowledge Fountain. For more information, please contact [email protected]. Insights into halophilic microbial adaptation: Analysis of integrons and associated genomic structures and characterization of a nitrilase in hypersaline environments School of Sciences and Engineering A Thesis Submitted by Sarah Ali Ahmed Sonbol. MSc to the Applied Sciences Graduate Program Spring, 2021 In partial fulfillment of the requirements for the degree of Doctorate in Applied Sciences (Biotechnology) Under the supervision of: Prof. -
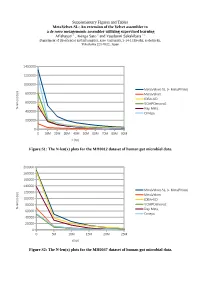
Supplementary Figures and Tables Metavelvet-SL
Supplementary Figures and Tables MetaVelvet-SL: An extension of the Velvet assembler to a de novo metagenomic assembler utilizing supervised learning Afiahayati 1 , Kengo Sato 1 and Yasubumi Sakakibara 1∗ Department of Biosciences and Informatics, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan 1400000 1200000 1000000 MetaVelvet-SL (+ MetaPhlAn) ) p 800000 b MetaVelvet ( ) x ( IDBA-UD n 600000 e SOAPDenovo2 l - N Ray Meta 400000 Omega 200000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S1: The N-len(x) plots for the MH0012 dataset of human gut microbial data. 200000 180000 160000 140000 MetaVelvet-SL (+ MetaPhlAn) ) 120000 p b MetaVelvet ( ) 100000 x IDBA-UD ( n e l 80000 SOAPDenovo2 - N Ray Meta 60000 Omega 40000 20000 0 0 5M 10M 15M 20M 25M x(bp) Figure S2: The N-len(x) plots for the MH0047 dataset of human gut microbial data. 600000 500000 400000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 300000 x IDBA-UD ( n e l SOAPDenovo2 - N 200000 Ray Meta Omega 100000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S3: The N-len(x) plots for the SRS017227 dataset of human gut microbial data. 800000 700000 600000 500000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 400000 x IDBA-UD ( n e l SOAPDenovo2 - 300000 N Ray Meta 200000 Omega 100000 0 0 5M 10M 15M 20M 25M 30M 35M 40M 45M x (bp) Figure S4: The N-len(x) plots for the SRS018661 dataset of human gut microbial data. Formula to identify unique nodes in Velvet (Zerbino and Birney, 2008) ̄x2 ρ2− log2 2 F ( ̄x ,n,ρ )= +n 2 2 where ̄x = the coverage of node ρ = expected coverage of subgraph n = the length of node A node is “unique”, if its F > 5 . -

Reorganising the Order Bacillales Through Phylogenomics
Systematic and Applied Microbiology 42 (2019) 178–189 Contents lists available at ScienceDirect Systematic and Applied Microbiology jou rnal homepage: http://www.elsevier.com/locate/syapm Reorganising the order Bacillales through phylogenomics a,∗ b c Pieter De Maayer , Habibu Aliyu , Don A. Cowan a School of Molecular & Cell Biology, Faculty of Science, University of the Witwatersrand, South Africa b Technical Biology, Institute of Process Engineering in Life Sciences, Karlsruhe Institute of Technology, Germany c Centre for Microbial Ecology and Genomics, University of Pretoria, South Africa a r t i c l e i n f o a b s t r a c t Article history: Bacterial classification at higher taxonomic ranks such as the order and family levels is currently reliant Received 7 August 2018 on phylogenetic analysis of 16S rRNA and the presence of shared phenotypic characteristics. However, Received in revised form these may not be reflective of the true genotypic and phenotypic relationships of taxa. This is evident in 21 September 2018 the order Bacillales, members of which are defined as aerobic, spore-forming and rod-shaped bacteria. Accepted 18 October 2018 However, some taxa are anaerobic, asporogenic and coccoid. 16S rRNA gene phylogeny is also unable to elucidate the taxonomic positions of several families incertae sedis within this order. Whole genome- Keywords: based phylogenetic approaches may provide a more accurate means to resolve higher taxonomic levels. A Bacillales Lactobacillales suite of phylogenomic approaches were applied to re-evaluate the taxonomy of 80 representative taxa of Bacillaceae eight families (and six family incertae sedis taxa) within the order Bacillales. -

Assessing Community Dynamics and Colonization Patterns Of
University of Vermont ScholarWorks @ UVM Graduate College Dissertations and Theses Dissertations and Theses 2017 Assessing Community Dynamics and Colonization Patterns of Tritatoma dimidiata and Other Biotic Factors Associated with Chagas Disease Prevalence in Central America Lucia Consuelo Orantes University of Vermont Follow this and additional works at: https://scholarworks.uvm.edu/graddis Part of the Genetics and Genomics Commons, and the Public Health Commons Recommended Citation Orantes, Lucia Consuelo, "Assessing Community Dynamics and Colonization Patterns of Tritatoma dimidiata and Other Biotic Factors Associated with Chagas Disease Prevalence in Central America" (2017). Graduate College Dissertations and Theses. 769. https://scholarworks.uvm.edu/graddis/769 This Dissertation is brought to you for free and open access by the Dissertations and Theses at ScholarWorks @ UVM. It has been accepted for inclusion in Graduate College Dissertations and Theses by an authorized administrator of ScholarWorks @ UVM. For more information, please contact [email protected]. ASSESSING COMMUNITY DYNAMICS AND COLONIZATION PATTERNS OF TRIATOMA DIMIDIATA AND OTHER BIOTIC FACTORS ASSOCIATED WITH CHAGAS DISEASE PREVALENCE IN CENTRAL AMERICA A Dissertation Presented by Lucia C. Orantes to The Faculty of the Graduate College of The University of Vermont In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Specializing in Natural Resources October, 2017 Defense Date: April 19, 2017 Dissertation Examination Committee: Kimberly Wallin, Ph.D., Advisor Sara Helms Cahan Ph.D., Advisor Donna Rizzo, Ph.D., Chairperson Leslie Morrissey, Ph.D. Gillian Galford, Ph.D. Cynthia J. Forehand, Ph.D., Dean of the Graduate Colleg ABSTRACT Chagas disease is caused by the parasite Trypanosoma cruzi and transmitted by multiple triatomine vectors across the Americas. -

Anaerovirgula Multivorans Gen. Nov., Sp. Nov., a Novel Spore-Forming, Alkaliphilic Anaerobe Isolated from Owens Lake, California, USA
International Journal of Systematic and Evolutionary Microbiology (2006), 56, 2623–2629 DOI 10.1099/ijs.0.64198-0 Anaerovirgula multivorans gen. nov., sp. nov., a novel spore-forming, alkaliphilic anaerobe isolated from Owens Lake, California, USA Elena V. Pikuta,1 Takashi Itoh,2 Paul Krader,3 Jane Tang,4 William B. Whitman5 and Richard B. Hoover1 Correspondence 1National Space Sciences and Technology Center/NASA, XD-12, 320 Sparkman Dr., Elena V. Pikuta Astrobiology Laboratory, Huntsville, AL 35805, USA [email protected] 2Japan Collection of Microorganisms, RIKEN BioResource Center, 2-1 Hirosawa, Wako-shi, Saitama 351-0198, Japan Richard B. Hoover 3 [email protected] American Type Culture Collection, 10801 University Blvd, Manassas, VA 20110, USA 4United States Department of Agriculture, Monitoring Programs Office, 8609 Sudley Rd, suite 206, Manassas, VA 20110, USA 5Department of Microbiology, University of Georgia, Athens, GA 30602-2605, USA A novel, alkaliphilic, obligately anaerobic bacterium, strain SCAT, was isolated from mud sediments of a soda lake in California, USA. The rod-shaped cells were motile, Gram-positive, formed spores and were 0?4–0?562?5–5?0 mm in size. Growth occurred within the pH range 6?7–10?0 and was optimal at pH 8?5. The temperature range for growth was 10–45 6C, with optimal growth at 35 6C. NaCl was required for growth. Growth occurred at 0?5–9?0 % (w/v) NaCl and was optimal at 1–2 % (w/v). The novel isolate was a catalase-negative chemo-organoheterotroph that fermented sugars, proteolysis products, some organic and amino acids, glycerol, D-cellobiose and cellulose. -

Analysis of the Structure of the Bacterial Community in the Livestock Manure-Based Composting Process*
113 Asian-Aust. J. Anim. Sci. Vol. 22, No. 1 : 113 - 118 January 2009 www.ajas.info Analysis of the Structure of the Bacterial Community in the Livestock Manure-based Composting Process* Hiraku Sasaki, Jun Nonaka, Kenichi Otawa, Osamu Kitazume, Ryoki Asano Takako Sasaki and Yutaka Nakai** Graduate School of Agricultural Science, Tohoku University, Aoba-ku, Sendai-shi, 981-8555, Japan ABSTRACT : We investigated the structure of bacterial communities present in livestock manure-based composting processes and evaluated the bacterial succession during the composting processes. Compost samples were derived separately from swine manure, dairy manure and sewage sludge. The structure of the bacterial community was analyzed by polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) using universal eubacterial primers. The genus Bacillus and related genera were mainly detected following the thermophilic composting phase of swine and dairy manure composts, and the members of the phylum Bacteroidetes were mainly detected in the cattle manure waste-based and sewage sludge compost. We recovered and sequenced limited number of the bands; however, the PCR-DGGE analysis showed that predominant diversities during the composting processes were markedly changed. Although PCR-DGGE analysis revealed the presence of different phyla in the early stages of composting, the members of the phylum Firmicutes and Bacteroidetes were observed to be one of the predominant phyla after the thermophilic phase. (Key Words : Bacterial Community, Compost, Denaturing Gradient Gel Electrophoresis, Manure) INTRODUCTION technique is the polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) method that can In livestock manure treatment systems, various species differentiate PCR products of identical lengths differing in of bacteria play an important role in the decomposition of sequence by even a single base (Muyzer et al., 1993). -

Kido Einlpoeto Aalbe:AIO(W H
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (43) International Publication Date (10) International Publication Number 18 May 2007 (18.05.2007) PCT WO 2007/056463 A3 (51) International Patent Classification: AT, AU, AZ, BA, BB, BU, BR, BW, BY, BZ, CA, CL CN, C12P 19/34 (2006.01) CO, CR, CU, CZ, DE, DK, DM, DZ, EC, FE, EU, ES, H, GB, GD, GE, GIL GM, UT, IAN, HIR, HlU, ID, IL, IN, IS, (21) International Application Number: JP, KE, KG, KM, KN, Kg KR, KZ, LA, LC, LK, LR, LS, PCT/US2006/043502 LI, LU, LV, LY, MA, MD, MG, MK, MN, MW, MX, MY, M, PG, P, PL, PT, RO, RS, (22) International Filing Date:NA, NG, , NO, NZ, (22 InterntionaFilin Date:.006 RU, SC, SD, SE, SG, SK, SL, SM, SV, SY, TJ, TM, TN, 9NR, TI, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of regional protection available): ARIPO (BW, GIL GM, KE, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, ZM, (30) Priority Data: ZW), Eurasian (AM, AZ, BY, KU, KZ, MD, RU, TJ, TM), 60/735,085 9 November 2005 (09.11.2005) US European (AT, BE, BU, CIL CY, CZ, DE, DK, EE, ES, H, FR, GB, UR, IJU, JE, IS, IT, LI, LU, LV, MC, NL, PL, PT, (71) Applicant (for all designated States except US): RO, SE, SI, SK, IR), GAPI (BE BJ, C, CU, CI, CM, GA, PRIMERA BIOSYSTEMS, INC. -

Temproal Change in Soil Microbiome Under Influence of Eight Years of Manure Amendment in Cultivated Soils
TEMPROAL CHANGE IN SOIL MICROBIOME UNDER INFLUENCE OF EIGHT YEARS OF MANURE AMENDMENT IN CULTIVATED SOILS By XUWEN WIENEKE Bachelor of Science in Animal Science Northwest A&F University Yangling, Shaanxi 2009 Master of Science in Animal Genetics and Breeding South Dakota State University Brookings, SD 2013 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY December, 2017 TEMPROAL CHANGE IN SOIL MICROBIOME UNDER INFLUENCE OF EIGHT YEARS OF MANURE AMENDMENT IN CULTIVATED SOILS Dissertation Approved: Dr. Udaya Desilva Dissertation Adviser Dr. Andrew Doust Dr. Michael Anderson Dr. Scott Carter Dr. Stephen Clarke ii ACKNOWLEDGEMENTS I dedicate this dissertation to our Lord, Jesus Christ. All the achievements in our lives are because of His blessings. I could not wait to show my appreciation to my adviser, Dr. Udaya Desilva. Udaya took me in as a refugee and has truly been my mentor. He earns my full respect and trust by being himself-wise, kind, caring, honest, and trustworthy. Having him as the role model, I have been learning on how to do research and how to be there for others. Life has never been better. I full-heartedly thank my committee members, Dr. Andrew Doust, Dr. Michael Anderson, Dr. Scott Carter, and Dr. Stephen Clarke. You are wise and have always been patient and supportive. I look up to you all and appreciate your tremendous helps. Many thanks to Dr. Noha Youssef. She has been helping me so much on my projects. My appreciation goes to Dr.