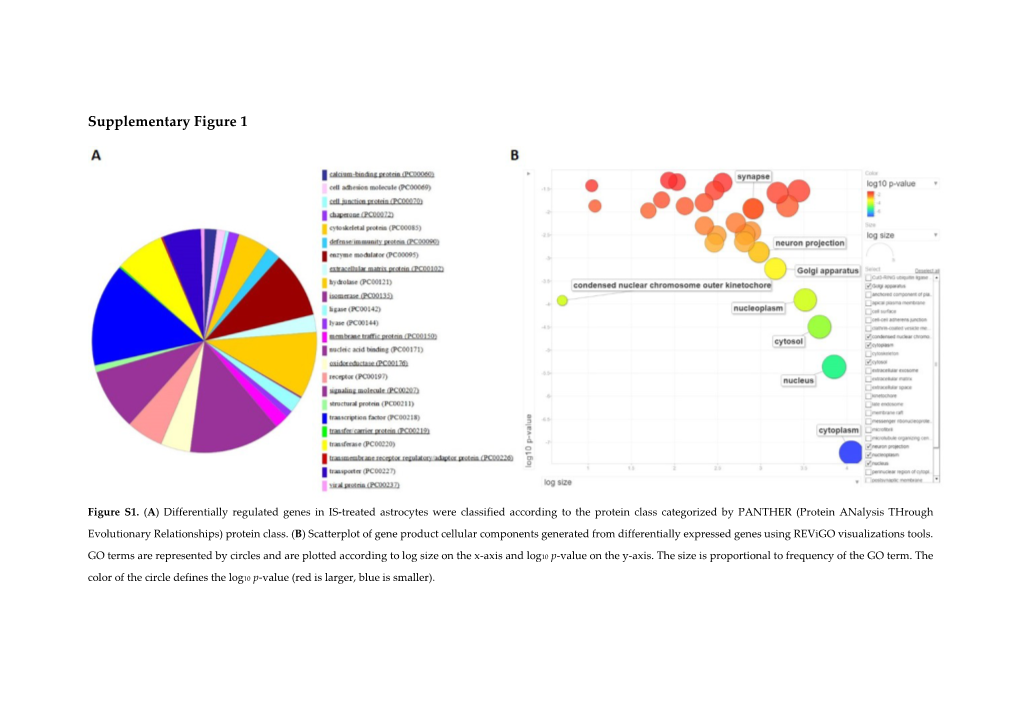
Supplementary Figure 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deregulated Gene Expression Pathways in Myelodysplastic Syndrome Hematopoietic Stem Cells
Leukemia (2010) 24, 756–764 & 2010 Macmillan Publishers Limited All rights reserved 0887-6924/10 $32.00 www.nature.com/leu ORIGINAL ARTICLE Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells A Pellagatti1, M Cazzola2, A Giagounidis3, J Perry1, L Malcovati2, MG Della Porta2,MJa¨dersten4, S Killick5, A Verma6, CJ Norbury7, E Hellstro¨m-Lindberg4, JS Wainscoat1 and J Boultwood1 1LRF Molecular Haematology Unit, NDCLS, John Radcliffe Hospital, Oxford, UK; 2Department of Hematology Oncology, University of Pavia Medical School, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy; 3Medizinische Klinik II, St Johannes Hospital, Duisburg, Germany; 4Division of Hematology, Department of Medicine, Karolinska Institutet, Stockholm, Sweden; 5Department of Haematology, Royal Bournemouth Hospital, Bournemouth, UK; 6Albert Einstein College of Medicine, Bronx, NY, USA and 7Sir William Dunn School of Pathology, University of Oxford, Oxford, UK To gain insight into the molecular pathogenesis of the the World Health Organization.6,7 Patients with refractory myelodysplastic syndromes (MDS), we performed global gene anemia (RA) with or without ringed sideroblasts, according to expression profiling and pathway analysis on the hemato- poietic stem cells (HSC) of 183 MDS patients as compared with the the French–American–British classification, were subdivided HSC of 17 healthy controls. The most significantly deregulated based on the presence or absence of multilineage dysplasia. In pathways in MDS include interferon signaling, thrombopoietin addition, patients with RA with excess blasts (RAEB) were signaling and the Wnt pathways. Among the most signifi- subdivided into two categories, RAEB1 and RAEB2, based on the cantly deregulated gene pathways in early MDS are immuno- percentage of bone marrow blasts. -

Deletion of Endoplasmic Reticulum Stress-Responsive Co-Chaperone P58ipk Protects Mice from Diet-Induced Steatohepatitis
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2018 Deletion of endoplasmic reticulum stress-responsive co-chaperone p58IPK protects mice from diet-induced steatohepatitis Bandla, Harikrishna ; Dasgupta, Debanjali ; Mauer, Amy S ; Nozickova, Barbora ; Kumar, Swarup ; Hirsova, Petra ; Graham, Rondell P ; Malhi, Harmeet DOI: https://doi.org/10.1111/hepr.13052 Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-144890 Journal Article Accepted Version Originally published at: Bandla, Harikrishna; Dasgupta, Debanjali; Mauer, Amy S; Nozickova, Barbora; Kumar, Swarup; Hirsova, Petra; Graham, Rondell P; Malhi, Harmeet (2018). Deletion of endoplasmic reticulum stress-responsive co-chaperone p58IPK protects mice from diet-induced steatohepatitis. Hepatology Research, 48(6):479- 494. DOI: https://doi.org/10.1111/hepr.13052 Deletion of endoplasmic reticulum stress-responsive co-chaperone p58IPK protects mice from diet-induced steatohepatitis Harikrishna Bandla1, Debanjali Dasgupta1, Amy S. Mauer1, Barbora Nozickova2, Swarup Kumar3, Petra Hirsova1, Rondell P. Graham4, Harmeet Malhi1* 1. Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN 2. Universitatsspital Zurich, 8096, Ramistrasse 100, Zurich, Switzerland 3. Department of Medicine, Saint Vincent Hospital, 123 Summer St, Worcester, MA 4. Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN Corresponding author: Harmeet Malhi, M.B.B.S. Associate Professor of Medicine and Physiology Mayo Clinic College of Medicine 200 First Street SW Rochester, MN 55905 Tel: 507 284 0686 Fax: 507 284 0762 Email: [email protected] Funding: This work was supported by DK 97178, DK107402 and DK111378 (H.M.), the Robert and Elizabeth Strickland Career Development Award from the Division of Endocrinology (H.M.), the Gilead Sciences Research Scholars Program in Liver Disease (H.M.) and the Palumbo Foundation (H.M.), the Edward C. -
![Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]](https://docslib.b-cdn.net/cover/8283/computational-genome-wide-identification-of-heat-shock-protein-genes-in-the-bovine-genome-version-1-peer-review-2-approved-1-approved-with-reservations-88283.webp)
Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]
F1000Research 2018, 7:1504 Last updated: 08 AUG 2021 RESEARCH ARTICLE Computational genome-wide identification of heat shock protein genes in the bovine genome [version 1; peer review: 2 approved, 1 approved with reservations] Oyeyemi O. Ajayi1,2, Sunday O. Peters3, Marcos De Donato2,4, Sunday O. Sowande5, Fidalis D.N. Mujibi6, Olanrewaju B. Morenikeji2,7, Bolaji N. Thomas 8, Matthew A. Adeleke 9, Ikhide G. Imumorin2,10,11 1Department of Animal Breeding and Genetics, Federal University of Agriculture, Abeokuta, Nigeria 2International Programs, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, 14853, USA 3Department of Animal Science, Berry College, Mount Berry, GA, 30149, USA 4Departamento Regional de Bioingenierias, Tecnologico de Monterrey, Escuela de Ingenieria y Ciencias, Queretaro, Mexico 5Department of Animal Production and Health, Federal University of Agriculture, Abeokuta, Nigeria 6Usomi Limited, Nairobi, Kenya 7Department of Animal Production and Health, Federal University of Technology, Akure, Nigeria 8Department of Biomedical Sciences, Rochester Institute of Technology, Rochester, NY, 14623, USA 9School of Life Sciences, University of KwaZulu-Natal, Durban, 4000, South Africa 10School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, 30032, USA 11African Institute of Bioscience Research and Training, Ibadan, Nigeria v1 First published: 20 Sep 2018, 7:1504 Open Peer Review https://doi.org/10.12688/f1000research.16058.1 Latest published: 20 Sep 2018, 7:1504 https://doi.org/10.12688/f1000research.16058.1 Reviewer Status Invited Reviewers Abstract Background: Heat shock proteins (HSPs) are molecular chaperones 1 2 3 known to bind and sequester client proteins under stress. Methods: To identify and better understand some of these proteins, version 1 we carried out a computational genome-wide survey of the bovine 20 Sep 2018 report report report genome. -

C-Terminal HSP90 Inhibitors Block the HSP90:HIF-1Α Interaction and Inhibit the Cellular Hypoxic Response
bioRxiv preprint doi: https://doi.org/10.1101/521989; this version posted January 24, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. C-terminal HSP90 Inhibitors Block the HSP90:HIF-1α Interaction and Inhibit the Cellular Hypoxic Response Nalin Katariaa, Bernadette Kerra, Samantha S. Zaiterb, Shelli McAlpineb and Kristina M Cooka† a. University of Sydney, Faculty of Medicine and Health, Charles Perkins Centre, Sydney, Australia. b. School of Chemistry, University of New South Wales, Sydney, Australia. † To whom correspondence should be addressed: Kristina M Cook, Charles Perkins Centre, University of Sydney, Sydney, NSW, Australia 2006; [email protected]; Tel: +61 286274858. Hypoxia Inducible Factor (HIF) is a transcription factor cancer cells known as a heat shock response (HSR)12. The activated by low oxygen, which is common in solid compounds also have poor selectivity for HSP9013,14. tumours. HIF controls the expression of genes involved in C-terminus inhibitors of HSP90 (SM molecules)11 act in a angiogenesis, chemotherapy resistance and metastasis. The selective manner, and in contrast to the N-terminus chaperone HSP90 (Heat Shock Protein 90) stabilizes the inhibitors, they do not induce a heat shock response13,15. subunit HIF-1α and prevents degradation. Previously Although the SM molecules block all co-chaperones that identified HSP90 inhibitors bind to the N-terminal pocket bind to the C-terminus of HSP90 and inhibit HSP90 of HSP90 which blocks binding to HIF-1α, and produces function16,17, there is no data discussing whether C- HIF-1α degradation. -

Defining Functional Interactions During Biogenesis of Epithelial Junctions
ARTICLE Received 11 Dec 2015 | Accepted 13 Oct 2016 | Published 6 Dec 2016 | Updated 5 Jan 2017 DOI: 10.1038/ncomms13542 OPEN Defining functional interactions during biogenesis of epithelial junctions J.C. Erasmus1,*, S. Bruche1,*,w, L. Pizarro1,2,*, N. Maimari1,3,*, T. Poggioli1,w, C. Tomlinson4,J.Lees5, I. Zalivina1,w, A. Wheeler1,w, A. Alberts6, A. Russo2 & V.M.M. Braga1 In spite of extensive recent progress, a comprehensive understanding of how actin cytoskeleton remodelling supports stable junctions remains to be established. Here we design a platform that integrates actin functions with optimized phenotypic clustering and identify new cytoskeletal proteins, their functional hierarchy and pathways that modulate E-cadherin adhesion. Depletion of EEF1A, an actin bundling protein, increases E-cadherin levels at junctions without a corresponding reinforcement of cell–cell contacts. This unexpected result reflects a more dynamic and mobile junctional actin in EEF1A-depleted cells. A partner for EEF1A in cadherin contact maintenance is the formin DIAPH2, which interacts with EEF1A. In contrast, depletion of either the endocytic regulator TRIP10 or the Rho GTPase activator VAV2 reduces E-cadherin levels at junctions. TRIP10 binds to and requires VAV2 function for its junctional localization. Overall, we present new conceptual insights on junction stabilization, which integrate known and novel pathways with impact for epithelial morphogenesis, homeostasis and diseases. 1 National Heart and Lung Institute, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. 2 Computing Department, Imperial College London, London SW7 2AZ, UK. 3 Bioengineering Department, Faculty of Engineering, Imperial College London, London SW7 2AZ, UK. 4 Department of Surgery & Cancer, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Endoplasmic Reticulum Chaperone Erdj4 Is Required for Survival, Glucose Metabolism and B Cell Development
The endoplasmic reticulum chaperone ERdj4 is required for survival, glucose metabolism and B cell development A dissertation submitted to the University of Cincinnati in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in the Immunobiology Graduate Program of the College of Medicine 2012 by Jill Marie Fritz B.A., Miami University, Oxford, OH Advisory Committee: Timothy Weaver, M.S., Ph.D., Chair George Deepe, M.D. Fred Finkelman, M.D. H. Leighton Grimes, Ph.D. Christopher Karp, M.D. Francis McCormack, M.D. ABSTRACT The ER-localized DnaJ homologue 4 (ERdj4) is a soluble ER chaperone induced by the unfolded protein response (UPR) to assist in the removal of unfolded/misfolded proteins from the ER lumen for proteasomal degradation. To elucidate the function of ERdj4 in vivo, ERdj4 gene trap (ERdj4GT/GT) mice were generated from embryonic stem cells harboring a gene trap cassette inserted into the ERdj4 locus. ERdj4GT/GT mice expressed hypomorphic levels of ERdj4 with a 10-100 fold reduction in all tissues and cell types examined. Approximately 30-50% of ERdj4GT/GT mice died perinatally in association with growth retardation and hypoglycemia. ERdj4GT/GT neonates exhibited signs of delayed pancreatic development, including abnormal distribution of pancreatic α- and β-cells and reduced insulin and glucagon in the pancreas. Surviving adult ERdj4GT/GT mice were glucose intolerant, resulting from hypoinsulinemia rather than insulin resistance. Pancreatic β-cells exhibited increased ER stress, including ER dilation and upregulation of the UPR. Proinsulin accumulated in the ER of β-cells from ERdj4GT/GT mice consistent with our previous finding that proinsulin is a substrate for ERdj4. -

Gene Expression Analysis of Angioimmunoblastic Lymphoma Indicates Derivation from T Follicular Helper Cells and Vascular Endothelial Growth Factor Deregulation
Research Article Gene Expression Analysis of Angioimmunoblastic Lymphoma Indicates Derivation from T Follicular Helper Cells and Vascular Endothelial Growth Factor Deregulation Pier Paolo Piccaluga,1,2 Claudio Agostinelli,1 Andrea Califano,3 Antonino Carbone,4 Luca Fantoni,6 Sergio Ferrari,5 Anna Gazzola,1 Annunziata Gloghini,6 Simona Righi,1 Maura Rossi,1 Enrico Tagliafico,5 Pier Luigi Zinzani,1 Simonetta Zupo,7 Michele Baccarani,1 and Stefano A. Pileri1 1Institute of Hematology and Medical Oncology ‘‘L. and A. Sera`gnoli,’’ S. Orsola-Malpighi Hospital, University of Bologna, Bologna, Italy; 2Institute for Cancer Genetics and 3Center for Computational Biology and Biochemistry, Columbia University, New York, New York; 4Department of Pathology, Istituto Nazionale Tumori, Milan, Italy; 5Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy; 6Division of Experimental Oncology, Centro di Riferimento Oncologico, Aviano, Italy; and 7S.S.D. Diagnostica Malattie Linfoproliferative, Istituto Nazionale per la Ricerca sul Cancro, Genova, Italy Abstract Introduction Angioimmunoblastic lymphoma (AILT) is the second most Peripheral T-cell lymphomas (PTCL) represent 10% to 15% of common subtype of peripheral T-cell lymphoma (PTCL) and all lymphoid neoplasms (1). They are a heterogeneous group of is characterized by dismal prognosis. Thus far, only a fewstu- tumors that in the Revised European-American Lymphoma dies have dealt with its molecular pathogenesis. We performed (REAL)/WHO classification are subdivided into -

The UVB-Induced Gene Expression Profile of Human Epidermis in Vivo Is Different from That of Cultured Keratinocytes
Oncogene (2006) 25, 2601–2614 & 2006 Nature Publishing Group All rights reserved 0950-9232/06 $30.00 www.nature.com/onc ORIGINAL ARTICLE The UVB-induced gene expression profile of human epidermis in vivo is different from that of cultured keratinocytes CD Enk1, J Jacob-Hirsch2, H Gal3, I Verbovetski4, N Amariglio2, D Mevorach4, A Ingber1, D Givol3, G Rechavi2 and M Hochberg1 1Department of Dermatology, The Hadassah-Hebrew University Medical Center, Jerusalem, Israel; 2Department of Pediatric Hemato-Oncology and Functional Genomics, Safra Children’s Hospital, Sheba Medical Center and Sackler School of Medicine, Tel-Aviv University,Tel Aviv, Israel; 3Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel and 4The Laboratory for Cellular and Molecular Immunology, Department of Medicine, The Hadassah-Hebrew University Medical Center, Jerusalem, Israel In order to obtain a comprehensive picture of the radiation. UVB, with a wavelength range between 290 molecular events regulating cutaneous photodamage of and 320 nm, represents one of the most important intact human epidermis, suction blister roofs obtained environmental hazards affectinghuman skin (Hahn after a single dose of in vivo ultraviolet (UV)B exposure and Weinberg, 2002). To protect itself against the were used for microarray profiling. We found a changed DNA-damaging effects of sunlight, the skin disposes expression of 619 genes. Half of the UVB-regulated genes over highly complicated cellular programs, including had returned to pre-exposure baseline levels at 72 h, cell-cycle arrest, DNA repair and apoptosis (Brash et al., underscoring the transient character of the molecular 1996). Failure in selected elements of these defensive cutaneous UVB response. -

The Eif2 Phosphatase: Characterization and Modulation
The eIF2 phosphatase: characterization and modulation Ana Crespillo Casado Darwin College July 2018 Ana Crespillo Casado The eIF2 phosphatase: characterization and modulation Abstract Cellular needs are fulfilled by the combined activity of functional proteins. Consequently, cells are equipped with a complex proteostatic network that controls protein production in response to cellular requisites. Thereby, protein synthesis is induced or attenuated depending on the particular cellular conditions. One of the mechanisms to control protein synthesis is the phosphorylation of eIF2, which triggers the so-called Integrated Stress Response (ISR). Kinases that sense stresses induce the phosphorylation of eIF2, which, on the one hand, attenuates global rates of protein synthesis and, on the other hand, activates the expression of specific proteins that help to alleviate the stress. One of the proteins preferentially expressed during the ISR is PPP1R15A, a regulatory subunit of Protein Phosphatase 1 (PP1). The PP1/PPP1R15A holophosphatase dephosphorylates eIF2 and terminates the ISR once the stresses are resolved. Hence, eIF2 kinases and phosphatases work together to control levels of phosphorylated eIF2. Maintaining the right balance between the activity of these kinases and phosphatases is important, as is seen by the correlation between their perturbance and the appearance of certain cellular malfunctions or diseases. However, affecting this balance has been also suggested to have beneficial effects. For example, genetic interference with the PPP1R15A regulatory subunit is proposed to confer protection to mice and cells under ER-stress conditions. This observation led to the search for compounds with the ability to modulate the ISR, in particular, by acting on the eIF2 phosphatases. -

At Elevated Temperatures, Heat Shock Protein Genes Show Altered Ratios Of
EXPERIMENTAL AND THERAPEUTIC MEDICINE 22: 900, 2021 At elevated temperatures, heat shock protein genes show altered ratios of different RNAs and expression of new RNAs, including several novel HSPB1 mRNAs encoding HSP27 protein isoforms XIA GAO1,2, KEYIN ZHANG1,2, HAIYAN ZHOU3, LUCAS ZELLMER4, CHENGFU YUAN5, HAI HUANG6 and DEZHONG JOSHUA LIAO2,6 1Department of Pathology, Guizhou Medical University Hospital; 2Key Lab of Endemic and Ethnic Diseases of The Ministry of Education of China in Guizhou Medical University; 3Clinical Research Center, Guizhou Medical University Hospital, Guiyang, Guizhou 550004, P.R. China; 4Masonic Cancer Center, University of Minnesota, Minneapolis, MN 55455, USA; 5Department of Biochemistry, China Three Gorges University, Yichang, Hubei 443002; 6Center for Clinical Laboratories, Guizhou Medical University Hospital, Guiyang, Guizhou 550004, P.R. China Received December 16, 2020; Accepted May 10, 2021 DOI: 10.3892/etm.2021.10332 Abstract. Heat shock proteins (HSP) serve as chaperones genes may engender multiple protein isoforms. These results to maintain the physiological conformation and function of collectively suggested that, besides increasing their expres‑ numerous cellular proteins when the ambient temperature is sion, certain HSP and associated genes also use alternative increased. To determine how accurate the general assumption transcription start sites to produce multiple RNA transcripts that HSP gene expression is increased in febrile situations is, and use alternative splicing of a transcript to produce multiple the RNA levels of the HSF1 (heat shock transcription factor 1) mature RNAs, as important mechanisms for responding to an gene and certain HSP genes were determined in three cell increased ambient temperature in vitro. lines cultured at 37˚C or 39˚C for three days. -

Allele-Specific Silencing of Mutant Ataxin-7 in SCA7 Patient-Derived
European Journal of Human Genetics (2014) 22, 1369–1375 & 2014 Macmillan Publishers Limited All rights reserved 1018-4813/14 www.nature.com/ejhg ARTICLE Allele-specific silencing of mutant Ataxin-7 in SCA7 patient-derived fibroblasts Janine Scholefield1,3, Lauren Watson1,2,3, Danielle Smith2, Jacquie Greenberg2 and Matthew JA Wood*,1,2 Polyglutamine (polyQ) disorders are inherited neurodegenerative conditions defined by a common pathogenic CAG repeat expansion leading to a toxic gain-of-function of the mutant protein. Consequences of this toxicity include activation of heat- shock proteins (HSPs), impairment of the ubiquitin-proteasome pathway and transcriptional dysregulation. Several studies in animal models have shown that reducing levels of toxic protein using small RNAs would be an ideal therapeutic approach for such disorders, including spinocerebellar ataxia-7 (SCA7). However, testing such RNA interference (RNAi) effectors in genetically appropriate patient cell lines with a disease-relevant phenotype has yet to be explored. Here, we have used primary adult dermal fibroblasts from SCA7 patients and controls to assess the endogenous allele-specific silencing of ataxin-7 by two distinct siRNAs. We further identified altered expression of two disease-relevant transcripts in SCA7 patient cells: a twofold increase in levels of the HSP DNAJA1 and a twofold decrease in levels of the de-ubiquitinating enzyme, UCHL1. After siRNA treatment, the expression of both genes was restored towards normal levels. To our knowledge, this is the first time that allele- specific silencing of mutant ataxin-7, targeting a common SNP, has been demonstrated in patient cells. These findings highlight the advantage of an allele-specific RNAi-based therapeutic approach, and indicate the value of primary patient-derived cells as useful models for mechanistic studies and for measuring efficacy of RNAi effectors on a patient-to-patient basis in the polyQ diseases.