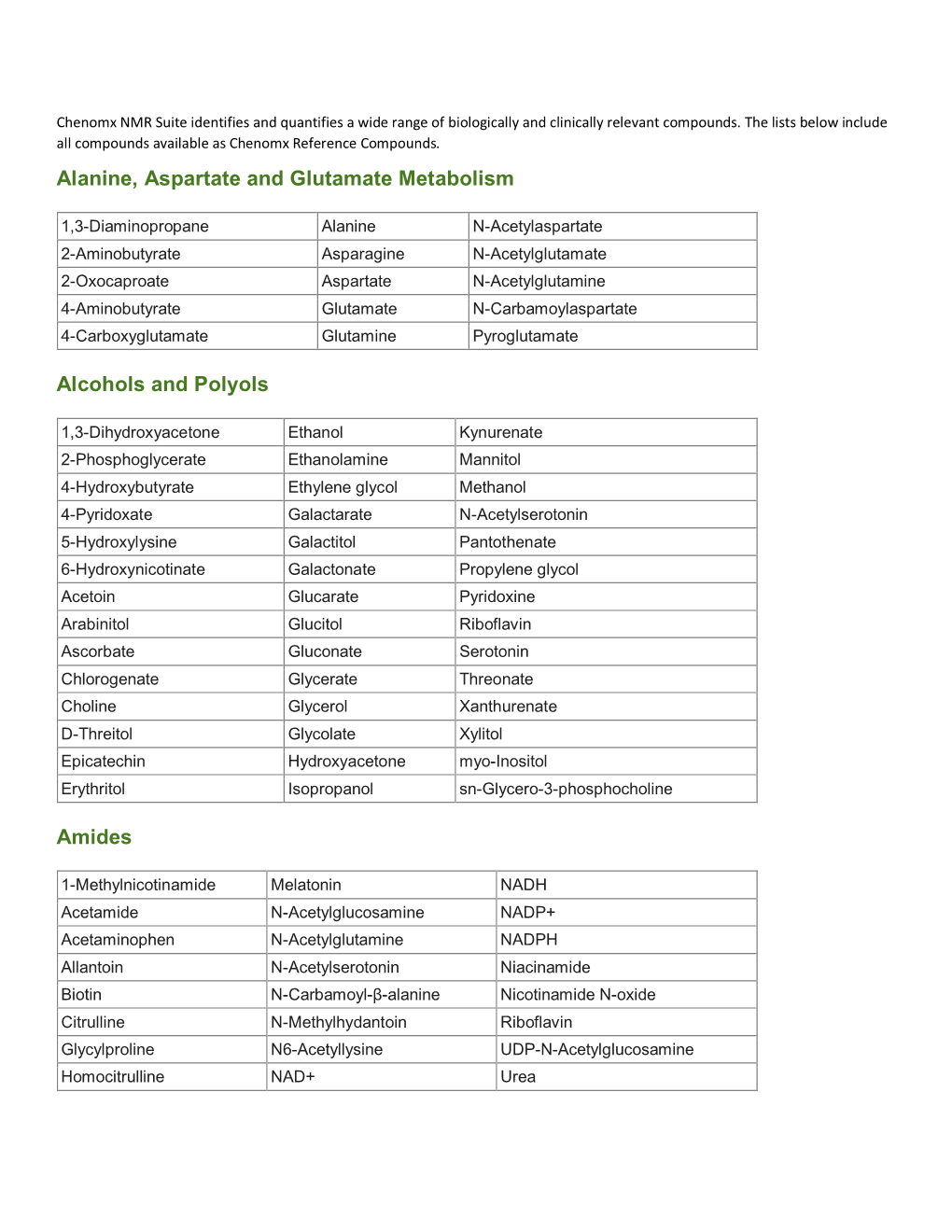
Alanine, Aspartate and Glutamate Metabolism Alcohols and Polyols
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Measurement of Metabolite Variations and Analysis of Related Gene Expression in Chinese Liquorice (Glycyrrhiza Uralensis) Plants
www.nature.com/scientificreports OPEN Measurement of metabolite variations and analysis of related gene expression in Chinese liquorice Received: 15 March 2017 Accepted: 28 March 2018 (Glycyrrhiza uralensis) plants under Published: xx xx xxxx UV-B irradiation Xiao Zhang1,2, Xiaoli Ding3,4, Yaxi Ji1,2, Shouchuang Wang5, Yingying Chen1,2, Jie Luo5, Yingbai Shen1,2 & Li Peng3,4 Plants respond to UV-B irradiation (280–315 nm wavelength) via elaborate metabolic regulatory mechanisms that help them adapt to this stress. To investigate the metabolic response of the medicinal herb Chinese liquorice (Glycyrrhiza uralensis) to UV-B irradiation, we performed liquid chromatography tandem mass spectrometry (LC-MS/MS)-based metabolomic analysis, combined with analysis of diferentially expressed genes in the leaves of plants exposed to UV-B irradiation at various time points. Fifty-four metabolites, primarily amino acids and favonoids, exhibited changes in levels after the UV-B treatment. The amino acid metabolism was altered by UV-B irradiation: the Asp family pathway was activated and closely correlated to Glu. Some amino acids appeared to be converted into antioxidants such as γ-aminobutyric acid and glutathione. Hierarchical clustering analysis revealed that various favonoids with characteristic groups were induced by UV-B. In particular, the levels of some ortho- dihydroxylated B-ring favonoids, which might function as scavengers of reactive oxygen species, increased in response to UV-B treatment. In general, unigenes encoding key enzymes involved in amino acid metabolism and favonoid biosynthesis were upregulated by UV-B irradiation. These fndings lay the foundation for further analysis of the mechanism underlying the response of G. -

Part One Amino Acids As Building Blocks
Part One Amino Acids as Building Blocks Amino Acids, Peptides and Proteins in Organic Chemistry. Vol.3 – Building Blocks, Catalysis and Coupling Chemistry. Edited by Andrew B. Hughes Copyright Ó 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim ISBN: 978-3-527-32102-5 j3 1 Amino Acid Biosynthesis Emily J. Parker and Andrew J. Pratt 1.1 Introduction The ribosomal synthesis of proteins utilizes a family of 20 a-amino acids that are universally coded by the translation machinery; in addition, two further a-amino acids, selenocysteine and pyrrolysine, are now believed to be incorporated into proteins via ribosomal synthesis in some organisms. More than 300 other amino acid residues have been identified in proteins, but most are of restricted distribution and produced via post-translational modification of the ubiquitous protein amino acids [1]. The ribosomally encoded a-amino acids described here ultimately derive from a-keto acids by a process corresponding to reductive amination. The most important biosynthetic distinction relates to whether appropriate carbon skeletons are pre-existing in basic metabolism or whether they have to be synthesized de novo and this division underpins the structure of this chapter. There are a small number of a-keto acids ubiquitously found in core metabolism, notably pyruvate (and a related 3-phosphoglycerate derivative from glycolysis), together with two components of the tricarboxylic acid cycle (TCA), oxaloacetate and a-ketoglutarate (a-KG). These building blocks ultimately provide the carbon skeletons for unbranched a-amino acids of three, four, and five carbons, respectively. a-Amino acids with shorter (glycine) or longer (lysine and pyrrolysine) straight chains are made by alternative pathways depending on the available raw materials. -

Expression and Regulation of the Rate-Limiting Enzymes of the Kynurenine Pathway in the Mouse Brain by Alexandra Kelly Brooks Di
EXPRESSION AND REGULATION OF THE RATE-LIMITING ENZYMES OF THE KYNURENINE PATHWAY IN THE MOUSE BRAIN BY ALEXANDRA KELLY BROOKS DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Neuroscience in the Graduate College of the University of Illinois at Urbana-Champaign, 2017 Urbana, Illinois Doctoral Committee: Research Assistant Professor Robert H. McCusker, Chair Assistant Professor Andrew J. Steelman Assistant Professor Monica Uddin Professor Rodney W. Johnson P a g e | ii ABSTRACT During the mid-1900’s, early anti-depressants were developed to increase levels of catecholamines within the brain, leading to the theory that depression symptomology was a result of an imbalance of neurotransmitters within the brain. To date, the catecholamine theory of depression is still held to be the prevailing theory as selective serotonin reuptake inhibitors being the most commonly prescribed anti-depressants. However, the incidence of depression is still rising with a vast amount of patients failing to respond to current treatments, and with this knowledge, additional theories of the neurobiology of depression have arisen over the past 30 years. A leading theory is Kynurenine Pathway activation in relation to inflammation- or stress- induced depression-like behaviors. There has been a tremendous amount of data connecting activated immune system (or hypothalamic-pituitary-adrenal axis), increased kynurenine production and depression symptomology. Thus, understanding the factors that activate this -

Tryptophan and Kynurenine Enhances the Stemness and Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stromal Cells in Vitro and in Vivo
materials Article Tryptophan and Kynurenine Enhances the Stemness and Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stromal Cells In Vitro and In Vivo Hai Thanh Pham 1,2 , Mitsuaki Ono 3,*, Emilio Satoshi Hara 4,* , Ha Thi Thu Nguyen 1,2,3, Anh Tuan Dang 1,2,3, Hang Thuy Do 1,2,3, Taishi Komori 1, Ikue Tosa 1, Yuri Hazehara-Kunitomo 1,3, Yuya Yoshioka 1, Yasutaka Oida 1, Kentaro Akiyama 1 and Takuo Kuboki 1 1 Department of Oral Rehabilitation and Regenerative Medicine, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama 700-8558, Japan; [email protected] (H.T.P.); [email protected] (H.T.T.N.); [email protected] (A.T.D.); [email protected] (H.T.D.); [email protected] (T.K.); [email protected] (I.T.); [email protected] (Y.H.-K.); [email protected] (Y.Y.); [email protected] (Y.O.); [email protected] (K.A.); [email protected] (T.K.) 2 Faculty of Dentistry, Hai Phong University of Medicine and Pharmacy, Haiphong 04211, Vietnam 3 Department of Molecular Biology and Biochemistry, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama 700-8558, Japan 4 Department of Biomaterials, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama 700-8558, Japan * Correspondence: [email protected] (M.O.); [email protected] (E.S.H.); Tel.: +81-86-235-7127 (M.O.); +81-86-235-6667 (E.S.H.); Fax: +81-86-222-7768 (M.O.); +81-86-235-6669 (E.S.H.) Abstract: Aging tissues present a progressive decline in homeostasis and regenerative capacities, Citation: Pham, H.T.; Ono, M.; Hara, which has been associated with degenerative changes in tissue-specific stem cells and stem cell E.S.; Nguyen, H.T.T.; Dang, A.T.; Do, niches. -

Figure S1. Heat Map of R (Pearson's Correlation Coefficient)
Figure S1. Heat map of r (Pearson’s correlation coefficient) value among different samples including replicates. The color represented the r value. Figure S2. Distributions of accumulation profiles of lipids, nucleotides, and vitamins detected by widely-targeted UPLC-MC during four fruit developmental stages. The colors indicate the proportional content of each identified metabolites as determined by the average peak response area with R scale normalization. PS1, 2, 3, and 4 represents fruit samples collected at 27, 84, 125, 165 Days After Anthesis (DAA), respectively. Three independent replicates were performed for each stages. Figure S3. Differential metabolites of PS2 vs PS1 group in flavonoid biosynthesis pathway. Figure S4. Differential metabolites of PS2 vs PS1 group in phenylpropanoid biosynthesis pathway. Figure S5. Differential metabolites of PS3 vs PS2 group in flavonoid biosynthesis pathway. Figure S6. Differential metabolites of PS3 vs PS2 group in phenylpropanoid biosynthesis pathway. Figure S7. Differential metabolites of PS4 vs PS3 group in biosynthesis of phenylpropanoids pathway. Figure S8. Differential metabolites of PS2 vs PS1 group in flavonoid biosynthesis pathway and phenylpropanoid biosynthesis pathway combined with RNA-seq results. Table S1. A total of 462 detected metabolites in this study and their peak response areas along the developmental stages of apple fruit. mix0 mix0 mix0 Index Compounds Class PS1a PS1b PS1c PS2a PS2b PS2c PS3a PS3b PS3c PS4a PS4b PS4c ID 1 2 3 Alcohols and 5.25E 7.57E 5.27E 4.24E 5.20E -

THE ROLE of KYNURENINE, a TRYPTOPHAN METABOLITE THAT INCREASES with AGE, in MUSCLE ATROPHY and LIPID PEROXIDATION) by Helen Eliz
THE ROLE OF KYNURENINE, A TRYPTOPHAN METABOLITE THAT INCREASES WITH AGE, IN MUSCLE ATROPHY AND LIPID PEROXIDATION) By Helen Elizabeth Kaiser Submitted to the Faculty of the Graduate School of Augusta University in partial fulfillment of the Requirements of the Degree of (Doctor of Philosophy in Cellular Biology and Anatomy) April 2020 COPYRIGHT© 2020 by Helen Kaiser ACKNOWLEDGEMENTS I wish to express my deepest gratitude to my mentor Dr. Mark Hamrick. His support and advice were invaluable to my learning and development as a scientist. I would like to especially recognize Dr. Hamrick’s honesty to data, and kindness to everyone around him. It was an honor to work as his student, and to be a part of this project. I would like to thank my committee members, Drs. Carlos Isales, Meghan McGee-Lawrence, and Yutao Liu, for their continued support, guidance, and helpful suggestions. They have helped to shape and focus this project, and have augmented my experience as a student during the development of my dissertation. Additionally, I want to recognize and thank Dr. Sadanand Fulzele for sharing his wealth of knowledge in techniques, and always keeping his door open for student’s questions. Next, a most heartfelt thanks to my lab mates: Andrew Khayrullin, Bharati Mendhe, Ling Ruan and Emily Parker. This project has been a team effort, all of them have been fundamental to its success. I further want to show my appreciation for the rest of the department of Cellular Biology and Anatomy at Augusta University. The histology core members Donna Kumiski and Penny Roon for their direct help in sectioning, and contributions to this project. -

An Integrated Meta-Analysis of Peripheral Blood Metabolites and Biological Functions in Major Depressive Disorder
Molecular Psychiatry https://doi.org/10.1038/s41380-020-0645-4 ARTICLE An integrated meta-analysis of peripheral blood metabolites and biological functions in major depressive disorder 1,2,3 1,2,3 1,2,3 1,3 1,3 4,5 1,3 1,3 Juncai Pu ● Yiyun Liu ● Hanping Zhang ● Lu Tian ● Siwen Gui ● Yue Yu ● Xiang Chen ● Yue Chen ● 1,2,3 1,3 1,3 1,3 1,3 1,2,3 Lining Yang ● Yanqin Ran ● Xiaogang Zhong ● Shaohua Xu ● Xuemian Song ● Lanxiang Liu ● 1,2,3 1,3 1,2,3 Peng Zheng ● Haiyang Wang ● Peng Xie Received: 3 June 2019 / Revised: 24 December 2019 / Accepted: 10 January 2020 © The Author(s) 2020. This article is published with open access Abstract Major depressive disorder (MDD) is a serious mental illness, characterized by high morbidity, which has increased in recent decades. However, the molecular mechanisms underlying MDD remain unclear. Previous studies have identified altered metabolic profiles in peripheral tissues associated with MDD. Using curated metabolic characterization data from a large sample of MDD patients, we meta-analyzed the results of metabolites in peripheral blood. Pathway and network analyses were then performed to elucidate the biological themes within these altered metabolites. We identified 23 differentially 1234567890();,: 1234567890();,: expressed metabolites between MDD patients and controls from 46 studies. MDD patients were characterized by higher levels of asymmetric dimethylarginine, tyramine, 2-hydroxybutyric acid, phosphatidylcholine (32:1), and taurochenode- soxycholic acid and lower levels of L-acetylcarnitine, creatinine, L-asparagine, L-glutamine, linoleic acid, pyruvic acid, palmitoleic acid, L-serine, oleic acid, myo-inositol, dodecanoic acid, L-methionine, hypoxanthine, palmitic acid, L-tryptophan, kynurenic acid, taurine, and 25-hydroxyvitamin D compared with controls. -

COMPILATION of AMINO ACIDS, DRUGS, METABOLITES and OTHER COMPOUNDS in MASSTRAK AMINO ACID ANALYSIS SOLUTION Paula Hong, Kendon S
COMPILATION OF AMINO ACIDS, DRUGS, METABOLITES AND OTHER COMPOUNDS IN MASSTRAK AMINO ACID ANALYSIS SOLUTION Paula Hong, Kendon S. Graham, Alexandre Paccou, T homas E. Wheat and Diane M. Diehl INTRODUCTION LC conditions Physiological amino acid analysis is commonly performed to LC System: Waters ACQUITY UPLC® System with TUV monitor and study a wide variety of metabolic processes. A wide Column: MassTrak AAA Column 2.1 x 150 mm, 1.7 µm variety of drugs, foods, and metabolic intermediates that may Column Temp: 43 ˚C be present in biological fluids can appear as peaks in amino Flow Rate: 400 µL/min. acid analysis, therefore, it is important to be able to identify Mobile Phase A: MassTrak AAA Eluent A Concentrate, unknown compounds.1,2,3 The reproducibility and robustness of diluted 1:10 the MassTrak Amino Acid Analysis Solution make this method well Mobile Phase B: MassTrak AAA Eluent B suited to such a study as well.4 Weak Needle Wash: 5/95 Acetonitrile/Water Strong Needle Wash: 95/5 Acetonitrile/Water Gradient: MassTrak AAA Standard Gradient (as provided in kit) Detection: UV @ 260 nm Injection Volume: 1 µL EXPERIMENTAL Injection Mode: Partial Loop with Needle Overfill (PLNO) Compound sample preparation A library of compounds was assembled. Each compound was derivatized individually and spiked into the MassTrak™ AAA Solution Standard prior to chromatographic analysis. The elution RESULTS AND DISCUSSION position of each tested compound could be related to known amino acids. A wide variety of antibiotics, pharmaceutical compounds and metabolite by-products are found in biological fluids. The reten- 1. -

Impacts of Dietary Exposure to Pesticides on Faecal Microbiome
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448511; this version posted June 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Impacts of dietary exposure to pesticides on faecal microbiome 2 metabolism in adult twins 3 4 Robin Mesnage1, Ruth C E Bowyer2, Souleiman El Balkhi3, Franck Saint-Marcoux3, Arnaud 5 Gardere3, Quinten Raymond Ducarmon4, Anoecim Robecca Geelen4, Romy Daniëlle Zwittink4, 6 Dimitris Tsoukalas5, Evangelia Sarandi5, Efstathia I. Paramera6, Timothy Spector2, Claire J Steves2, 7 Michael N Antoniou1* 8 9 1 Gene Expression and Therapy Group, King's College London, Faculty of Life Sciences & Medicine, 10 Department of Medical and Molecular Genetics, Guy's Hospital, London, SE1 9RT, UK. 11 12 2 Department of Twin Research and Genetic Epidemiology, Kings College London, London, UK 13 14 3 Service de pharmacologie, toxicologie et pharmacovigilance, UF Toxicologie analytique environnementale 15 et santé au travail, CHU de Limoges, Limoges, France 16 17 4 Center for Microbiome Analyses and Therapeutics, Leiden University Medical Center, Leiden, The 18 Netherlands 19 20 5 Metabolomic Medicine Clinic, Health Clinics for Autoimmune and Chronic Diseases, 10674 Athens, 21 Greece 22 23 6 NEOLAB S.A., Medical laboratory, 125 Michalakopoulu Str., 11527 Athens, Greece 24 25 26 *Correspondence: [email protected] 27 bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448511; this version posted June 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Intestinal Phosphate Transport in Familial ~~~O~Hos~Haternicrickets
FAMILIAL HYPOPHOSPHATEMIC RICKETS 69 1 pine oxidoreductase. The deficiencies were confirmed in skin Glutaric aciduria: A "new" disorder of amino acid metabolism. Biochem. fibroblasts from two siblings with the disease and a third patient Med., 12: 12 (1975). 15. Hutzler, J., and Dancis. J.: Saccharopine cleavage by a dehydrogenase of human from an unrelated family. liver. Biochim. Biophys. Acta. 206: 205 (1970). REFERENCES AND NOTES 16. Hutzler. J., and Dancis. J.: Preparative synthesis of saccharopine. Biochim. Biophys. Acta, 222: 225 (1970). I. Ampola. M. G., Ampola. M., Mian, M., Shaw, W., Levy, H., Letsou, A,, and 17. Hutzler, J., and Dancis, J.: Lysine-ketoglutarate reductase in human tissues. Doyle. M.: In preparation. Biochim. Biophys. Acta, 377: 42 (1975). 2. Bowden, J. A., and Connelly, J. L.: Branched-chain a-keto acid metabolism. I. 18. Krooth, R. S.: Constitutive mutations and hereditary enzyme deficiencies in Isolation, purification and partial characterization of bovine liver a- mammalian cells. In: M. Harris and B. Thompson: Regulation of Gene ketoisocaproic:a-keto-@-methylvaleric acid dehydrogenase. J. Biol. Chem., Expression in Eukaryotic Cells, p. 115 (Fogarty International Center, Be- 243: 1 198 (1968). thesda, Md., 1973). 3. Cox. R. P.. Krauss, M. R., Balis, M. E., and Dancis, J.: Communication between 19. Lormans, S., and Lowenthal. A,: Amino adipic aciduria in an oligophrenic child. normal and enzyme deficient cells in tissue culture. Exp. Cell Res.. 74: 251 Clin. Chim. Acta. 57: 97 (1974). (1972). 20. Lowry. 0. H., Rosehrough, N. J.. Farr. A. L., and Randall, R. J.: Protein 4. Cox. R. P., and MacLeod, C. -

Urinary Metabolomics to Identify a Unique Biomarker Panel for Detecting Colorectal Cancer: a Multicenter Study
Published OnlineFirst May 31, 2019; DOI: 10.1158/1055-9965.EPI-18-1291 Research Article Cancer Epidemiology, Biomarkers Urinary Metabolomics to Identify a Unique & Prevention Biomarker Panel for Detecting Colorectal Cancer: A Multicenter Study Lu Deng1, Kathleen Ismond1,2, Zhengjun Liu3, Jeremy Constable4, Haili Wang2, Olusegun I. Alatise5, Martin R. Weiser4, T.P. Kingham4, and David Chang1,2 Abstract Background: Population-based screening programs are paring the metabolomic profiles from colorectal cancer versus credited with earlier colorectal cancer diagnoses and treat- controls. Multiple models were constructed leading to a good ment initiation, which reduce mortality rates and improve separation of colorectal cancer from controls. patient health outcomes. However, recommended screen- Results: A panel of 17 metabolites was identified as possible ing methods are unsatisfactory as they are invasive, are biomarkers for colorectal cancer. Using only two of the select- resource intensive, suffer from low uptake, or have poor ed metabolites, namely diacetylspermine and kynurenine, a diagnostic performance. Our goal was to identify a urine predictor for detecting colorectal cancer was developed with an metabolomic-based biomarker panel for the detection of AUC of 0.864, a specificity of 80.0%, and a sensitivity of colorectal cancer that has the potential for global popula- 80.0%. tion-based screening. Conclusions: We present a potentially "universal" metabo- Methods: Prospective urine samples were collected from lomic biomarker panel for colorectal cancer independent of study participants. Based upon colonoscopy and histopathol- cohort clinical features based on a North American popula- ogy results, 342 participants (colorectal cancer, 171; healthy tion. Further research is needed to confirm the utility of the controls, 171) from two study sites (Canada, United States) profile in a prospective, population-based colorectal cancer were included in the analyses. -

Amino Acid Degradation
BI/CH 422/622 OUTLINE: OUTLINE: Protein Degradation (Catabolism) Digestion Amino-Acid Degradation Inside of cells Protein turnover Dealing with the carbon Ubiquitin Fates of the 29 Activation-E1 Seven Families Conjugation-E2 nitrogen atoms in 20 1. ADENQ Ligation-E3 AA: Proteosome 2. RPH 9 ammonia oxidase Amino-Acid Degradation 18 transamination Ammonia 2 urea one-carbon metabolism free transamination-mechanism to know THF Urea Cycle – dealing with the nitrogen SAM 5 Steps Carbamoyl-phosphate synthetase 3. GSC Ornithine transcarbamylase PLP uses Arginino-succinate synthetase Arginino-succinase 4. MT – one carbon metabolism Arginase 5. FY – oxidase vs oxygenase Energetics Urea Bi-cycle 6. KW – Urea Cycle – dealing with the nitrogen 7. BCAA – VIL Feeding the Urea Cycle Glucose-Alanine Cycle Convergence with Fatty acid-odd chain Free Ammonia Overview Glutamine Glutamate dehydrogenase Overall energetics Amino Acid A. Concepts 1. ConvergentDegradation 2. ketogenic/glucogenic 3. Reactions seen before The SEVEN (7) Families B. Transaminase (A,D,E) / Deaminase (Q,N) Family C. Related to biosynthesis (R,P,H; C,G,S; M,T) 1.Glu Family a. Introduce oxidases/oxygenases b. Introduce one-carbon metabolism (1C) 2.Pyruvate Family a. PLP reactions 3. a-Ketobutyric Family (M,T) a. 1-C metabolism D. Dedicated 1. Aromatic Family (F,Y) a. oxidases/oxygenases 2. a-Ketoadipic Family (K,W) 3. Branched-chain Family (V,I,L) E. Convergence with Fatty Acids: propionyl-CoA 29 N 1 Amino Acid Degradation • Intermediates of the central metabolic pathway • Some amino acids result in more than one intermediate. • Ketogenic amino acids can be converted to ketone bodies.