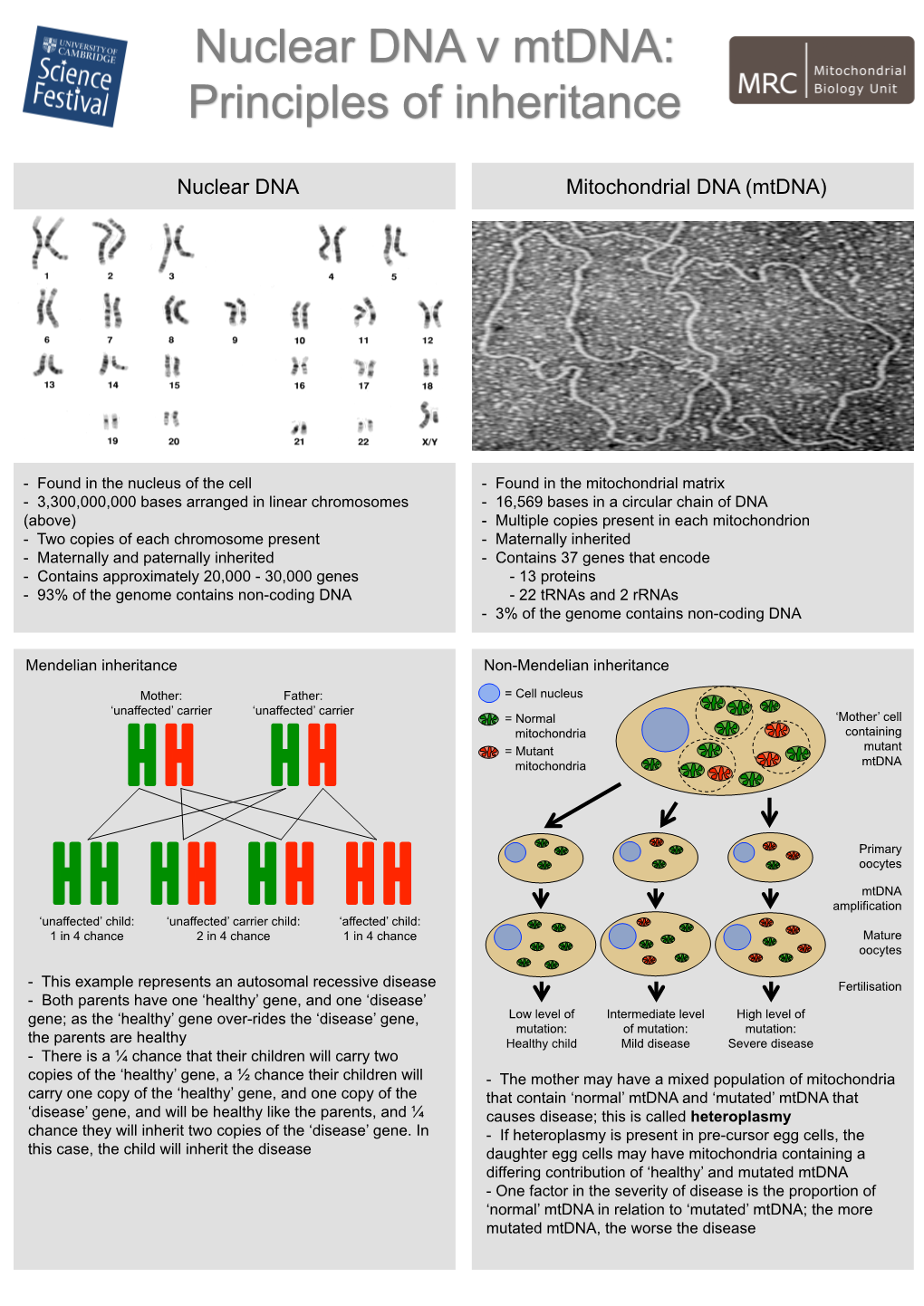
Nuclear DNA Mitochondrial DNA (Mtdna)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sequence-Selective Recognition of Double-Stranded RNA And
Downloaded from rnajournal.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Hnedzko et al. Sequence-Selective Recognition of Double-Stranded RNA and Enhanced Cellular Uptake of Cationic Nucleobase and Backbone- Modified Peptide Nucleic Acids Dziyana Hnedzko1,*, Dennis W. McGee,2 Yannis A. Karamitas1 and Eriks Rozners1,* Departments of 1 Chemistry and 2 Biological Sciences, Binghamton University, The State University of New York, Binghamton, New York 13902, United States. * To whom correspondence should be addressed. Tel: +1-607-777-2441; Fax: +1-607-777- 4478; Email: [email protected] and [email protected] Running Tittle: RNA recognition and cellular uptake of PNA 1 Downloaded from rnajournal.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Hnedzko et al. ABSTRACT Sequence-selective recognition of complex RNAs in live cells could find broad applications in biology, biomedical research and biotechnology. However, specific recognition of structured RNA is challenging and generally applicable and effective methods are lacking. Recently, we found that peptide nucleic acids (PNAs) were unusually well suited ligands for recognition of double-stranded RNAs. Herein, we report that 2-aminopyridine (M) modified PNAs and their conjugates with lysine and arginine tripeptides form strong (Ka = 9.4 to 17 × 107 M-1) and sequence-selective triple helices with RNA hairpins at physiological pH and salt concentration. The affinity of PNA-peptide conjugates for the matched RNA hairpins was unusually high compared to the much lower affinity for DNA hairpins of the same 7 -1 sequence (Ka = 0.05 to 0.11 × 10 M ). The binding of double-stranded RNA by M-modified 4 -1 -1 PNA-peptide conjugates was a relatively fast process (kon = 2.9 × 10 M s ) compared to 3 -1 -1 the notoriously slow triple helix formation by oligodeoxynucleotides (kon ~ 10 M s ). -

Mt-Atp8 Gene in the Conplastic Mouse Strain C57BL/6J-Mtfvb/NJ on the Mitochondrial Function and Consequent Alterations to Metabolic and Immunological Phenotypes
From the Lübeck Institute of Experimental Dermatology of the University of Lübeck Director: Prof. Dr. Saleh M. Ibrahim Interplay of mtDNA, metabolism and microbiota in the pathogenesis of AIBD Dissertation for Fulfillment of Requirements for the Doctoral Degree of the University of Lübeck from the Department of Natural Sciences Submitted by Paul Schilf from Rostock Lübeck, 2016 First referee: Prof. Dr. Saleh M. Ibrahim Second referee: Prof. Dr. Stephan Anemüller Chairman: Prof. Dr. Rainer Duden Date of oral examination: 30.03.2017 Approved for printing: Lübeck, 06.04.2017 Ich versichere, dass ich die Dissertation ohne fremde Hilfe angefertigt und keine anderen als die angegebenen Hilfsmittel verwendet habe. Weder vorher noch gleichzeitig habe ich andernorts einen Zulassungsantrag gestellt oder diese Dissertation vorgelegt. ABSTRACT Mitochondria are critical in the regulation of cellular metabolism and influence signaling processes and inflammatory responses. Mitochondrial DNA mutations and mitochondrial dysfunction are known to cause a wide range of pathological conditions and are associated with various immune diseases. The findings in this work describe the effect of a mutation in the mitochondrially encoded mt-Atp8 gene in the conplastic mouse strain C57BL/6J-mtFVB/NJ on the mitochondrial function and consequent alterations to metabolic and immunological phenotypes. This work provides insights into the mutation-induced cellular adaptations that influence the inflammatory milieu and shape pathological processes, in particular focusing on autoimmune bullous diseases, which have recently been reported to be associated with mtDNA polymorphisms in the human MT-ATP8 gene. The mt-Atp8 mutation diminishes the assembly of the ATP synthase complex into multimers and decreases mitochondrial respiration, affects generation of reactive oxygen species thus leading to a shift in the metabolic balance and reduction in the energy state of the cell as indicated by the ratio ATP to ADP. -

Mitochondrial Complex III Deficiency Associated with a Homozygous Mutation in UQCRQ
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector REPORT Mitochondrial Complex III Deficiency Associated with a Homozygous Mutation in UQCRQ Ortal Barel,1 Zamir Shorer,2 Hagit Flusser,2 Rivka Ofir,1 Ginat Narkis,1 Gal Finer,1 Hanah Shalev,2 Ahmad Nasasra,2 Ann Saada,3 and Ohad S. Birk1,4,* A consanguineous Israeli Bedouin kindred presented with an autosomal-recessive nonlethal phenotype of severe psychomotor retarda- tion and extrapyramidal signs, dystonia, athetosis and ataxia, mild axial hypotonia, and marked global dementia with defects in verbal and expressive communication skills. Metabolic workup was normal except for mildly elevated blood lactate levels. Brain magnetic resonance imaging (MRI) showed increased density in the putamen, with decreased density and size of the caudate and lentiform nuclei. Reduced activity specifically of mitochondrial complex III and variable decrease in complex I activity were evident in muscle biopsies. Homozygosity of affected individuals to UQCRB and to BCSIL, previously associated with isolated complex III deficiency, was ruled out. Genome-wide linkage analysis identified a homozygosity locus of approximately 9 cM on chromosome 5q31 that was further narrowed down to 2.14 cM, harboring 30 genes (logarithm of the odds [LOD] score 8.82 at q ¼ 0). All 30 genes were sequenced, revealing a single missense (p.Ser45Phe) mutation in UQCRQ (encoding ubiquinol-cytochrome c reductase, complex III subunit VII, 9.5 kDa), one of the ten nuclear -

Biomarkers of Mitotoxicity After Acute Liver Injury: Further Insights Into the Interpretation of Glutamate Dehydrogenase
Journal of Clinical and Translational Research 10.18053/Jctres/07.202101.005 MINI REVIEW Biomarkers of mitotoxicity after acute liver injury: further insights into the interpretation of glutamate dehydrogenase Mitchell R. McGill1,2* and Hartmut Jaeschke3 1. Department of Environmental Health Sciences, Fay W. Boozman College of Public Health, University of Arkansas for Medical Sciences, 4301 W. Markham St, Little Rock, AR, USA, 72205 2. Department of Pharmacology and Toxicology, College of Medicine, University of Arkansas for Medical Sciences, 4301 W. Markham St., Little Rock, AR, USA, 72205 3. Department of Pharmacology, Toxicology and Therapeutics, University of Kansas Medical Center, 3901 Rainbow Blvd., Kansas City, KS, USA, 66160 *Corresponding author Mitchell R. McGill, PhD Department of Environmental Health Sciences & Department of Pharmacology and Toxicology, University of Arkansas for Medical Sciences, Little Rock, AR Tel: +1 501-526-6696 Email: [email protected] Article information: Received: November 03, 2020 Revised: December 09, 2020 Accepted: December 10, 2020 Journal of Clinical and Translational Research 10.18053/Jctres/07.202101.005 ABSTRACT Background: Acetaminophen (APAP) is a popular analgesic, but overdose causes acute liver injury and sometimes death. Decades of research have revealed that mitochondrial damage is central in the mechanisms of toxicity in rodents, but we know much less about the role of mitochondria in humans. Due to the challenge of procuring liver tissue from APAP overdose patients, non-invasive mechanistic biomarkers are necessary to translate the mechanisms of APAP hepatotoxicity from rodents to patients. It was recently proposed that the mitochondrial matrix enzyme glutamate dehydrogenase (GLDH) can be measured in circulation as a biomarker of mitochondrial damage. -

Nuclear and Mitochondrial DNA Sequences from Two Denisovan Individuals
Nuclear and mitochondrial DNA sequences from two Denisovan individuals Susanna Sawyera,1, Gabriel Renauda,1, Bence Violab,c,d, Jean-Jacques Hublinc, Marie-Theres Gansaugea, Michael V. Shunkovd,e, Anatoly P. Dereviankod,f, Kay Prüfera, Janet Kelsoa, and Svante Pääboa,2 aDepartment of Evolutionary Genetics, Max Planck Institute for Evolutionary Anthropology, D-04103 Leipzig, Germany; bDepartment of Anthropology, University of Toronto, Toronto, ON M5S 2S2, Canada; cDepartment of Human Evolution, Max Planck Institute for Evolutionary Anthropology, D-04103 Leipzig, Germany; dInstitute of Archaeology and Ethnography, Russian Academy of Sciences, Novosibirsk, RU-630090, Russia; eNovosibirsk National Research State University, Novosibirsk, RU-630090, Russia; and fAltai State University, Barnaul, RU-656049, Russia Contributed by Svante Pääbo, October 13, 2015 (sent for review April 16, 2015; reviewed by Hendrik N. Poinar, Fred H. Smith, and Chris B. Stringer) Denisovans, a sister group of Neandertals, have been described on DNA to the ancestors of present-day populations across Asia the basis of a nuclear genome sequence from a finger phalanx and Oceania suggests that in addition to the Altai Mountains, (Denisova 3) found in Denisova Cave in the Altai Mountains. The they may have lived in other parts of Asia. In addition to the only other Denisovan specimen described to date is a molar (Deni- finger phalanx, a molar (Denisova 4) was found in the cave in sova 4) found at the same site. This tooth carries a mtDNA se- 2000. Although less than 0.2% of the DNA in the tooth derives quence similar to that of Denisova 3. Here we present nuclear from a hominin source, the mtDNA was sequenced and differed DNA sequences from Denisova 4 and a morphological description, from the finger phalanx mtDNA at only two positions, suggesting as well as mitochondrial and nuclear DNA sequence data, from it too may be from a Denisovan (2, 3). -

Tyramine and Amyloid Beta 42: a Toxic Synergy
biomedicines Article Tyramine and Amyloid Beta 42: A Toxic Synergy Sudip Dhakal and Ian Macreadie * School of Science, RMIT University, Bundoora, VIC 3083, Australia; [email protected] * Correspondence: [email protected]; Tel.: +61-3-9925-6627 Received: 5 May 2020; Accepted: 27 May 2020; Published: 30 May 2020 Abstract: Implicated in various diseases including Parkinson’s disease, Huntington’s disease, migraines, schizophrenia and increased blood pressure, tyramine plays a crucial role as a neurotransmitter in the synaptic cleft by reducing serotonergic and dopaminergic signaling through a trace amine-associated receptor (TAAR1). There appear to be no studies investigating a connection of tyramine to Alzheimer’s disease. This study aimed to examine whether tyramine could be involved in AD pathology by using Saccharomyces cerevisiae expressing Aβ42. S. cerevisiae cells producing native Aβ42 were treated with different concentrations of tyramine, and the production of reactive oxygen species (ROS) was evaluated using flow cytometric cell analysis. There was dose-dependent ROS generation in wild-type yeast cells with tyramine. In yeast producing Aβ42, ROS levels generated were significantly higher than in controls, suggesting a synergistic toxicity of Aβ42 and tyramine. The addition of exogenous reduced glutathione (GSH) was found to rescue the cells with increased ROS, indicating depletion of intracellular GSH due to tyramine and Aβ42. Additionally, tyramine inhibited the respiratory growth of yeast cells producing GFP-Aβ42, while there was no growth inhibition when cells were producing GFP. Tyramine was also demonstrated to cause increased mitochondrial DNA damage, resulting in the formation of petite mutants that lack respiratory function. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Ribozymes Targeted to the Mitochondria Using the 5S Ribosomal Rna
RIBOZYMES TARGETED TO THE MITOCHONDRIA USING THE 5S RIBOSOMAL RNA By JENNIFER ANN BONGORNO A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2005 Copyright 2005 by Jennifer Bongorno To my grandmother, Hazel Traster Miller, whose interest in genealogy sparked my interest in genetics, and without whose mitochondria I would not be here ACKNOWLEDGMENTS I would like to thank all the members of the Lewin lab; especially my mentor, Al Lewin. Al was always there for me with suggestions and keeping me motivated. He and the other members of the lab were like my second family; I would not have had an enjoyable experience without them. Diana Levinson and Elizabeth Bongorno worked with me on the fourth and third mouse transfections respectively. Joe Hartwich and Al Lewin tested some of the ribozymes in vitro and cloned some of the constructs I used. James Thomas also helped with cloning and was an invaluable lab manager. Verline Justilien worked on a related project and was a productive person with whom to bounce ideas back and forth. Lourdes Andino taught me how to use the new phosphorimager for my SYBR Green-stained gels. Alan White was there through it all, like the older brother I never had. Mary Ann Checkley was with me even longer than Alan, since we both came to Florida from Ohio Wesleyan, although she did manage to graduate before me. Jia Liu and Frederic Manfredsson were there when I needed a beer. -

Into the Mitochondrial and Nuclear DNA and RNA of Two Transplantable Hepatomas (3924A and H-35Tc2) and Host Livers'
[CANCERRESEARCH28.2164-2167,October1968] Brief Communication Comparative Incorporation of Tritiated Thymidine and Cytidine into the Mitochondrial and Nuclear DNA and RNA of Two Transplantable Hepatomas (3924A and H-35tc2) and Host Livers' Lillie 0. Chang, Harold P.Morris,2and William B. Looney Radiobiology and Biophysics Laboratory, Department of Radiology, University of Virginia School of Medicine, Charlottesville, Vir ginia f@29O1(L. 0. C. and W. B. L.), and Laboratory of Biochemistry, National Cancer institute, NIH, Bethesda, Maryland ?LXJ114 (H. P. M.) Summary interesting questions as to the different precursor pools and It has been found that the specific activity of thymidine-5- the biosynthetic pathways of nucleic acid synthesis in the methyl-3H incorporation into the mitochondrial DNA of two mitochondria and nuclei of the hepatomas as well as of their transplantable hepatomas (3924A and H-35tc2) and host livers host livers. The purpose of this communication is to report the is less than the specific activity of the nuclear DNA. However, relative rates of uptake of the labeled thymidine and cytidine the ratio of mitochondrial DNA to nuclear DNA specific ac into DNA, and the incorporation of cytidine into RNA of the tivity in both hepatomas and host livers is 10—100times greater mitochondrial and nuclear fractions of Hepatomas 3924A and H-35tc2 and their host livers. following cytidine-5-3H administration than the ratio following thymidine-5-methyl-3H administration. This marked difference in the ratios of the specific activities of thymidine and cytidine Materials and Methods in mitochondrial and nuclear DNA of hepatomas and host livers The growth patterns of transplantable rat hepatoma lines appears to be the result of both an increase in cytidine incor have been described by Morris (8, 9) . -

Isolation of Active Genes Containing CAG Repeats by DNA Strand Invasion by a Peptide Nucleic Acid (Chromatin/Transcription Factors/Microsatellite DNA) LIDIA C
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 1901-1905, March 1995 Biochemistry Isolation of active genes containing CAG repeats by DNA strand invasion by a peptide nucleic acid (chromatin/transcription factors/microsatellite DNA) LIDIA C. BOFFA*, ELISABETrA M. CARPANETO*, AND VINCENT G. ALLFREYtt *Istituto Nazionale per la Ricerca sul Cancro IST, Genoa 16132, Italy; and tThe Rockefeller University, New York, NY 10021 Communicated by Bruce Merrifield, The Rockefeller University, New York, NY December 7, 1994 ABSTRACT An amplification of tandem CAG trinucle- can be captured quantitatively on streptavidin-coated mag- otide sequences in DNA due to errors in DNA replication is netic beads. involved in at least four hereditary neurodegenerative dis- An amplification of CAG triplet repeats occurs in at least eases. The CAG triplet repeats when translated into protein four inherited neurodegenerative diseases. In all cases, the give rise to tracts of glutamine residues, which are a promi- severity of the disease increases with the expansion of the CAG nent feature of many transcription factors, including the repeat region that occurs as a consequence of errors in DNA TATA-binding protein of transcription factor TFIID. We have replication. In Huntington disease, expansion of the CAG used a biotin-labeled, complementary peptide nucleic acid repeats in the gene results in an elongation of a glutamine-rich (PNA) to invade the CAG repeats in intact chromatin and then domain in the encoded protein from 35 to over 100 residues employed a method for the selective isolation of transcrip- (11). The functional consequence of similar glutamine expan- tionally active chromatin restriction fragments containing the sions for transcriptional control is indicated by the finding that PNA-DNA hybrids. -

Non-Coding RNA: Microrna Stimulates Mitochondrial Translation
RESEARCH HIGHLIGHTS Nature Reviews Genetics | AOP, published online 12 August 2014 IN BRIEF THERAPEUTICS Targeting huntingtin through morpholino oligomers Huntington’s disease is caused by a mutant huntingtin (HTT) gene that contains an expanded tract of poly(CAG) repeats. Sun et al. designed phosphorodiamidate morpholino oligomers (PMOs, which are stable nucleic acid mimics) as antisense reagents to target the CAG tract in HTT mRNA. Applying the PMOs to HTT-mutant human neurons in vitro decreased HTT protein levels and reduced toxicity caused by mutant HTT. Furthermore, in two mouse models of Huntington’s disease, intracranial injection of PMOs resulted in downregulation of mutant HTT levels and partially reduced disease symptoms. ORIGINAL RESEARCH PAPER Sun, X. et al. Phosphorodiamidate morpholino oligomers suppress mutant huntingtin expression and attenuate neurotoxicity. Hum. Mol. Genet. http://dx.doi.org/10.1093/hmg/ddu349 (2014) TECHNOLOGY Using DNA repair to detect modified bases There is great interest in characterizing the locations and functions of chemically modified bases in genomes. Bryan et al. report their Excision-seq method, in which DNA repair enzymes are used to cut genomic DNA at sites of the particular damaged bases they recognize, followed by high-throughput sequencing to characterize these cleavage sites. The researchers characterized the locations and sequence contexts of uracils (that is, demethylated thymines) in Escherichia coli and Saccharomyces cerevisiae genomes, and of ultraviolet-light-induced pyrimidine dimers in S. cerevisiae. ORIGINAL RESEARCH PAPER Bryan, D. S. et al. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. http://dx.doi.org/10.1101/ gr.174052.114 (2014) NON-CODING RNA MicroRNA stimulates mitochondrial translation The muscle-specific microRNA miR‑1 stimulates translation of various transcripts encoded by mitochondrial DNA, while repressing its nuclear DNA-encoded targets in the cytoplasm, report Zhang and colleagues. -

Protection Against Apoptosis by Monoamine Oxidase a Inhibitors
View metadata,FEBS 20082 citation and similar papers at core.ac.uk FEBS Letters 426 (1998)brought to 155^159 you by CORE provided by Elsevier - Publisher Connector Protection against apoptosis by monoamine oxidase A inhibitors W. Malornia;*, A.M. Giammariolia, P. Matarresea, P. Pietrangelib, E. Agostinellib, A. Ciacciob, E. Grassillic, B. Mondovi'b aDepartment of Ultrastructures, Istituto Superiore di Sanitaé, Viale Regina Elena 299, 00161 Rome, Italy bDepartment of Biochemical Sciences and CNR Center of Molecular Biology, University of Rome `La Sapienza', Rome, Italy cDepartment of General Pathology, University of Modena, Modena, Italy Received 27 February 1998 mitochondrial membrane potential or apoptosis. Analytical Abstract Several lines of evidence have been accumulating indicating that an important role may be played by mitochondrial cytology analyses revealed that maintenance of the mitochon- homeostasis in the initiation phase, the first stage of apoptosis. drial homeostasis by pargyline and clorgyline is associated This work describes the results obtained by using different with a partial hindering of the apoptotic process. inhibitors of monoamine oxidases (MAO), i.e. pargyline, clorgyline and deprenyl, on mitochondrial integrity and apopto- 2. Materials and methods sis. Both pargyline and clorgyline are capable of protecting cells from apoptosis induced by serum starvation while deprenyl is 2.1. Cell cultures ineffective. These data represent the first demonstration that Human melanoma cells (M14) were grown in monolayer in modi- MAO-A inhibitors may protect cells from apoptosis through a ¢ed RPMI 1640 medium supplemented with 10% heat-inactivated mechanism involving the maintenance of mitochondrial homeo- fetal calf serum (FCS), 1 mM sodium pyruvate, 1% non-essential stasis.