Bacterial and Viral Source Tracking in the Pocantico and Sparkill Creek Watersheds
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bobcats in Westchester County
Michael Rubbo, Ph.D. Department of Environmental Studies and Science Pace University 50 mi N of NYC Population of ~ 1 million . 44th most populated county in US 450 mi2 . 290,000 acres Significant open space . Over 100,ooo acres ▪ ~50% forested Approximately 9,500 acres Located in: . Towns of Ossining, Mount Pleasant, New Castle . Villages of Briarcliff Manor, Sleepy Hollow, Pleasantville Pocantico River originates in Echo Lake and ends at Hudson River . Flows north to south . Approximately 9.5 miles in length Sources: Esri, HERE, DeLorme, Intermap, increment P Corp., GEBCO, USGS, FAO, NPS, NRCAN, GeoBase, IGN, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), swisstopo, MapmyIndia, © OpenStreetMap contributors, and the GIS User Community Public . Rockefeller Park and Preserve – Over 1,700 acres . Hardscrabble Wilderness Area - ~250 acres . Pocantico Lake County Park - ~165 acres Private – open to the public . Stone Barns Center for Food and Agriculture Private . Edith Macy Conference Center - ~400 acres . Campfire Club - ~225 acres Identify unique ecological attributes or areas that are impaired . Will direct preservation/restoration efforts . Serve as basis for watershed management plan First step is to locate resources . Habitats are a good representation of overall biological resources Collected common data layers: . Roads . Municipal tax parcels . Topographic contours . Bedrock geology . Surficial geology . Soils . Hydrography . DEC streams . FEMA floodplains . Department of Environmental Conservation (DEC) wetlands . National Wetlands Inventory (NWI) wetlands . NY Natural Heritage Program (NYNHP) data . Agricultural land Standard hydrography and wetlands data sets Also used soil properties for wetlands . Used soils classes of: ▪ Somewhat Poorly Drained ▪ Poorly Drained ▪ Very Poorly Drained Crest, Ledge, and Talus . -
Waterbody Classifications, Streams Based on Waterbody Classifications
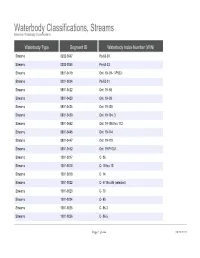
Waterbody Classifications, Streams Based on Waterbody Classifications Waterbody Type Segment ID Waterbody Index Number (WIN) Streams 0202-0047 Pa-63-30 Streams 0202-0048 Pa-63-33 Streams 0801-0419 Ont 19- 94- 1-P922- Streams 0201-0034 Pa-53-21 Streams 0801-0422 Ont 19- 98 Streams 0801-0423 Ont 19- 99 Streams 0801-0424 Ont 19-103 Streams 0801-0429 Ont 19-104- 3 Streams 0801-0442 Ont 19-105 thru 112 Streams 0801-0445 Ont 19-114 Streams 0801-0447 Ont 19-119 Streams 0801-0452 Ont 19-P1007- Streams 1001-0017 C- 86 Streams 1001-0018 C- 5 thru 13 Streams 1001-0019 C- 14 Streams 1001-0022 C- 57 thru 95 (selected) Streams 1001-0023 C- 73 Streams 1001-0024 C- 80 Streams 1001-0025 C- 86-3 Streams 1001-0026 C- 86-5 Page 1 of 464 09/28/2021 Waterbody Classifications, Streams Based on Waterbody Classifications Name Description Clear Creek and tribs entire stream and tribs Mud Creek and tribs entire stream and tribs Tribs to Long Lake total length of all tribs to lake Little Valley Creek, Upper, and tribs stream and tribs, above Elkdale Kents Creek and tribs entire stream and tribs Crystal Creek, Upper, and tribs stream and tribs, above Forestport Alder Creek and tribs entire stream and tribs Bear Creek and tribs entire stream and tribs Minor Tribs to Kayuta Lake total length of select tribs to the lake Little Black Creek, Upper, and tribs stream and tribs, above Wheelertown Twin Lakes Stream and tribs entire stream and tribs Tribs to North Lake total length of all tribs to lake Mill Brook and minor tribs entire stream and selected tribs Riley Brook -

Hudson River National Estuarine Research Reserve Management Plan OCTOBER 1, 2019–OCTOBER 1, 2024
Hudson River National Estuarine Research Reserve Management Plan OCTOBER 1, 2019–OCTOBER 1, 2024 Andrew M. Cuomo, Governor | Basil Seggos, Commissioner Acknowledgments This plan was prepared by staff of the Hudson River National Estuarine Research Reserve, including Betsy Blair, Chris Bowser, Ann-Marie Caprioli, Brian DeGasperis, Sarah Fernald, Heather Gierloff, Emilie Hauser, Dan Miller, and Sarah Mount, with the assistance of Andy Burgher, Cathy Kittle, and Bill Rudge in the New York State Department of Environmental Conservation; Ed McGowan of the New York State Office of Parks, Recreation and Historic Preservation; and Nina Garfield and Ann Weaver of the National Oceanic and Atmospheric Administration, Office for Coastal Management. We appreciate input that has informed development of this plan provided by other colleagues, local leaders, county officials, environmental organizations, researchers, educators, and marsh managers. Suggested citation: New York State Department of Environmental Conservation (NYSDEC). 2019. Hudson River National Estuarine Research Reserve Management Plan. Albany, NY. Table of Contents Executive Summary ................................................................................................................................... iv Introduction ................................................................................................................................................. 1 The Reserve ......................................................................................................................................... -

How's the Water in the Catskill, Esopus and Rondout Creeks?
How’s the Water in the Catskill, Esopus and Rondout Creeks? Cizen Science Fecal Contaminaon Study How’s the Water in the Catskill, Esopus and Rondout Creeks? Background & Problem Methods Results: 2012-2013 Potenal Polluon Sources © Riverkeeper 2014 © Riverkeeper 2014 Photo: Rob Friedman “SWIMMABILITY” FECAL PATHOGEN CONTAMINATION LOAD © Riverkeeper 2014 Government Pathogen Tesng © Riverkeeper 2014 Riverkeeper’s Fecal Contaminaon Study 2006 - Present Enterococcus (“Entero”) EPA-recommended fecal indicator Monthly sampling: May – Oct EPA Guideline for Primary Contact: Acceptable: 0-60 Entero per 100 mL Beach Advisory: >60 Entero per 100 mL © Riverkeeper 2014 Science Partners & Supporters Funders Science Partners • HSBC • Dr. Gregory O’Mullan Queens • Clinton Global Iniave College, City University of New • The Eppley Foundaon for York Research • Dr. Andrew Juhl, Lamont- • The Dextra Baldwin Doherty Earth Observatory, McGonagle Foundaon, Inc. Columbia University • The Hudson River Foundaon for Science and Environmental Research, Inc. • Hudson River Estuary Program, NYS DEC • New England Interstate Water Polluon Control Commission (2008-2013) © Riverkeeper 2014 Riverkeeper’s Cizen Science Program Goals 1. Fill a data gap 2. Raise awareness about fecal contaminaon in tributaries 3. Involve local residents in finding and eliminang Photo: John Gephards sources of contaminaon © Riverkeeper 2014 Riverkeeper’s Cizen Science Studies Tributaries sampled: • Catskill Creek • 45 river miles • 19 sites (many added in 2014) • Esopus Creek • 25 river miles -

Rockefeller Park Trail Map.Pdf
400 300 250 350 250 350 300 300 300 d R 300 rd 350 fo ed )¥ EA B 350 [k 350 µ 400 300 OCA 300 d 350 a RBR o r R 250 w e 300 o 350 l l EA 450 350 Ï[ o tillm e S a H n v k y L a p n i LO RBR LL L e TRAIL MAP le T o S S P c L 400 R i a d Rockwood t k l @@300 n o e O Rd Wa ic UP t y a t an c n R Rockefeller TB r o a d 300 G c LL n s P K ing o 300 P 400 350 o State Park Preserve DL Map produced by NYS OPRHP GIS Bureau, May 29, 2014. MI 150 Rockwood Hall Preserve Entrance YÉ s DL PR RR LL d FO 350 PP 150 600 u OCA DR 550 Legend BR 150 300 300 EA 150 DR 650 state park preserve H barn/farm BR 350 550 250 200 TB BP RR DL BH FO Spook Rock 350 other park land bridge LO RI LL 700 650 PR e BR 150 150 SH k Rockefeller Lands building a OV L OC NW RF 150 n Stone Barns Center a LR dam 200 RF BH wBP RI TB _) 400 S RF parking areas Trail ID NAME BT [k GE RI 500 farm _) d 450 wetlands Glacial BP R AS - Ash Tree Loop 200 TB Erratic WS 500 d gate PR RF r PW SR GB 200 waterbody 250 o RF BT - Big Tree Loop RF f LR EH RF d BR - Brook Trail 200 e park office EH 350 streams SH OV BP B 250 FM BP - Brothers' Path BT AS 350 OCA 10' contours 200 parking lot BH - Buttermilk Hill 300 Stone Barns Center 150 FL CA - Canter Alley RF minor roads 50 EH for WS RF DL - David's Loop d FM Food and Agriculture picnic site a RF FL o major roads DR - Deer Run R AS RF 200 GB RF restroom EH - Eagle Hill Trail w 300 350 state parkway )¥ 200 o GO PR PR l RF RF EA - Equestrian Access Trail l FS o PE rock formation PR H trails FM - Farm Meadow Trail50 BT y RF Sleepy PW p RF visitor center -

2.4 Natural Resources
EXISTING CONDITIONS Natural Environment 2.4. NATURAL RESOURCES foundations may result in structural failures or higher construction and maintenance costs. Briarcliff Manor’s image and character is shaped as much by the high quality of its natural setting as by its built environment. The Village’s Soils are classified by the Natural Resource Conservation Service into character is created by large privately owned open space parcels and four Hydrologic Soil Groups (HSG) based on the soil's runoff potential. golf courses, parkland, brooks, streams and the Hudson River and an The four Hydrologic Soils Groups are A, B, C and D; A’s generally abundance of trees. The protection of these features is essential to the have the smallest runoff potential and D’s the greatest. Soils in the preservation of Briarcliff Manor’s unique community character. Village are shown in Figure 2-7. The characteristics of the Village’s predominant soil types by HSG are described below: 2.4.1 Natural Resource Features Group A is sand, loamy sand or sandy loam types of soils. It has low runoff potential and high infiltration rates even when thoroughly Significant environmental systems and features are steep slopes, wetted. This group consists chiefly of deep, well to excessively drained floodplains and freshwater wetlands, fragile soils, and other prominent sands or gravels and has a high rate of water transmission. Little of the natural features. Each of these features is and should be a unique soil in the Village is of this type. Woodbridge loam (WdA) is located in community resource. five small scattered areas of the Village: the area around Dalmeny Road, two locations in the Chilmark neighborhood west of Old Topography Briarcliff Road, and vertical bands in the central and east central parts of the Village. -

Eastern NY Excluding Long Island 2014
DISCONTINUED SURFACE-WATER DISCHARGE OR STAGE-ONLY STATIONS The following continuous-record surface-water discharge or stage-only stations (gaging stations) in eastern New York excluding Long Island have been discontinued. Daily streamflow or stage records were collected and published for the period of record, expressed in water years, shown for each station. Those stations with an asterisk (*) before the station number are currently operated as crest-stage partial-record station and those with a double asterisk (**) after the station name had revisions published after the site was discontinued. [Letters after station name designate type of data collected: (d) discharge, (e) elevation, (g) gage height] Period of Station Drainage record Station name number area (mi2) (water years) HOUSATONIC RIVER BASIN Tenmile River near Wassaic, NY (d) 01199420 120 1959-61 Swamp River near Dover Plains, NY (d) 01199490 46.6 1961-68 Tenmile River at Dover Plains, NY (d) 01199500 189 1901-04 BLIND BROOK BASIN Blind Brook at Rye, NY (d) 01300000 8.86 1944-89 BEAVER SWAMP BROOK BASIN Beaver Swamp Brook at Mamaroneck, NY (d) 01300500 4.42 1944-89 MAMARONECK RIVER BASIN Mamaroneck River at Mamaroneck, NY (d) 01301000 23.1 1944-89 HUTCHINSON RIVER BASIN Hutchinson River at Pelham, NY (d) 01301500 6.04 1944-89 BRONX RIVER BASIN Bronx River at Bronxville, NY (d) 01302000 26.5 1944-89 HUDSON RIVER BASIN Opalescent River near Tahawus, NY (d) 01311900 9.02 1921-23 Arbutus Pond Outlet near Newcomb, NY (d) *01311992 1.22 1991-92 Cedar River near Indian Lake, NY (d) -

Village of Piermont Flood Guide
Know Your Risk and Stay Informed! Village of Piermont Know Your Zone: If you live close to the Sparkill Creek or the Flood Preparedness Guide for Residents and Businesses Hudson River, you may live in the flood zone and you should be pre- pared to secure your home and/or business and evacuate quickly. If One of the wonderful things about living in the Village of Piermont you’re not sure if you are at risk, check the map in this guide. Flood is being near the water. The Hudson River and Sparkill Creek are part of maps are also available through the Piermont Building Department what makes the Village such a special place. But, the river and the creek www.piermont-ny.gov/building-department or at the Piermont Vil- put some areas of the Village at risk of flooding. lage Hall at 478 Piermont Ave. Flooding is not just a problem for people who live near the water. Sign Up for CodeRed: Rockland County residents can sign up for All residents should be aware of risks and know how to find information CodeRed to receive notifications for emergency situations. and help when flooding occurs. Whether you live in the flood zone or not, Register at public.coderedweb.com or contact the Rockland County being prepared for an emergency is important. Keep this guide handy so Department of Fire & Emergency Services at (845) 638-5070. you know where to find help if you need it. New York Alert: You can sign up for the New York State notifica- tion system to receive phone calls or emails that will alert you to seri- Produced with ous or emergency situations. -

Hudson River Studies with Guest Scientist Dr. Robin E. Bell
Reconstructing the Storm and Industrial History of the Hudson River with Dr. Dallas Abbott 11 Feb 2017 The Hudson at the base of the Palisades on which LDEO is located is one of America’s most interesting waterways Few rivers match its beauty and history Autumn foliage, Alpine Kearney House, Alpine In the 19th Century, the “Hudson River School” was among America’s earliest art movements. “Hastings-on-Hudson” by Jasper Cropsey (1897.) http://www.byu.edu/moa/exhibits/Current%20Exhibits/150years/830025900.html “View from the Artist’s Home, Sunset” by Jasper Cropsey (1887). http://www.newingtoncropsey.com/index1.htm The river is named for Henry Hudson Hudson, an English explorer working for the Dutch East India Company, sailed up the river in the “Half Moon” in 1609 He was seeking the “Northwest Passage” around North America http://www.ulster.net/~hrmm/halfmoon/1609moon.htm Replica “Half Moon” In honor of the 400th anniversary of Hudson’s voyage, a full size replica of the “Half Moon” has returned to the river http://www.halfmoon. mus.ny.us/ August 2011 Up until development of modern highways in the 20th century, the Hudson was a major north-south transportation corridor. Dutch fur traders and settlers used the river between Ft. Orange (Albany) and New Amsterdam (NYC) Hundreds of thousands of people and tons of freight floated on the river to and from the Erie Canal’s eastern end, then on to the West. http://www.ulster.net/~hrmm/sail/age.html Even today, there is much barge traffic for cement, petroleum, and other goods. -

Table of Contents
WEST MOUNTAIN, GLENS FALLS, NY. JAMES BLEECKER ANNUAL REPORT TABLE OF CONTENTS 2001 Messages from the President and Board Chair 1 A Victory for the Hyde Park Heritage Corridor and Regional Economy 2 Making Waves on the River in a Nationally Significant Cause 4 Turning Industrial Wastelands into Riverfront Gems 6 Continuing Leadership in Power Plant and Reindustrialization Issues 8 Closing the Barn Door on Sprawl by Saving Agricultural Landscapes 10 Land Preservation Milestones 12 Riverfront Communities Milestones 14 Environmental Quality Milestones 16 Communications & Public Outreach Milestones 18 Scenic Hudson Volunteers 20 Development Milestones 22 Scenic Hudson Supporters 24 Financial Overview 29 Staff 32 Board of Directors 33 ANNUAL REPORT2001 Messages from the PRESIDENT and BOARD CHAIR Those who contemplate the beauty of the Earth find reserves of strength that will endure as long as life lasts. There is symbolic as well as actual beauty in the migration of birds, the ebb and flow of tides, the folded bud ready for spring. There is something infinitely healing in the repeated refrains of nature – the assurance that dawn comes after the night and spring after the winter. Rachel Carson – biologist, writer and ecologist achel he new Carson’s millen- Rwords T nium has are as meaning- been met by a ful today as they powerful evolu- were at the tion at Scenic TOM LIGAMARI TOM LIGAMARI dawn of the Hudson. Our modern environmental movement, which she inspired. capacity to affect the future of the valley has doubled Scenic Hudson, Inc.’s efforts during the past year car- in two years, steadily accelerating our work in open ried forward a commitment to our mission and the spir- space preservation, riverfront community revitalization it of her work. -

Community Water Quality Monitoring Results Sparkill Creek 2011‐2016 Overview
COMMUNITY WATER QUALITY MONITORING RESULTS SPARKILL CREEK 2011‐2016 OVERVIEW Riverkeeper and the Sparkill Creek Watershed Alliance have been testing the water for the fecal- indicator bacteria Enterococcus (“Entero”) since 2011. Sources of fecal contamination may include sewage infrastructure failures, sewer overflows, inadequate sewage treatment, septic system failures, agricultural runoff, urban runoff, and wildlife. Samples were collected monthly (approx. May to October) at 18 watershed locations by SCWA members and processed by Riverkeeper (2011-2015) and the Sarah Lawrence Center for the Urban River at Beczak (2016). A total of 572 samples have been analyzed since 2011. This water quality monitoring study is designed to learn about broad trends. The data can help inform choices about recreation in the creek, but cannot predict future water quality at any particular time and place. To see all the results visit www.riverkeeper.org/water-quality/citizen-data/sparkill-creek. WATERSHED SNAPSHOT These results are for non‐tidal sites only. As measured against the EPA’s recommended geometric mean (a weighted average) criterion for As measured against the Environmental Protection safe swimming: Agency’s recommended Beach Action Value for safe swimming: EPA GM threshold Sparkill Creek GM 30 746.3 93% cells/100 mL cells/100 mL of Sparkill Creek samples failed. After rainy weather: 3 Best Sites 3 Worst Sites Blauvelt‐ Tackamack Tappan‐ Moturis at Greenbush Br (#4) (#13) 98% Orangeburg‐ Piermont‐ Skating of Sparkill Creek samples failed. -

How's the Water? Hudson River Water Quality and Water Infrastructure
HOW’S THE WATER? Hudson River Water Quality and Water Infrastructure The Hudson River Estuary is an engine of life for the coastal ecosystem, the source of drinking water for more than 100,000 people, home to the longest open water swim event in the world, and the central feature supporting the quality of life and $4.4 billion tourism economy for the region. This report focuses on one important aspect of protecting and improving Hudson River Estuary water quality – sewage-related contamination and water infrastructure. Untreated sewage puts drinking water and recreational users at risk. Water quality data presented here are based on analysis of more than 8,200 samples taken since 2008 from the Hudson River Estuary by Riverkeeper, CUNY Queens College, Columbia University’s Lamont- Doherty Earth Observatory; and from its tributaries by dozens of partner organizations and individual 21% community scientists. Water infrastructure information Hudson River Estuary samples presented here is based on data from the Department that failed to meet federal safe of Environmental Conservation and Environmental swimming guidelines Facilities Corporation, which administers State Revolving Funds. 44 Municipally owned wastewater While the Hudson River is safe for swimming at most treatment plants that locations on most days sampled, raw sewage overflows discharge to the Estuary and leaks from aging and failing infrastructure too often make waters unsafe. The Hudson’s tributaries $4.8 Billion – the smaller creeks and rivers that feed it – are often Investment needed in sources of contamination. wastewater infrastructure in the Hudson River Watershed To improve water quality, action is needed at the federal, state and local levels to increase and prioritize infrastructure investments.