Genome-Wide Association Study and Admixture Mapping Reveal New Loci Associated with Total Ige Levels in Latinos
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected]
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School July 2017 Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the Pathology Commons Scholar Commons Citation Mohamed, Mai, "Role of Amylase in Ovarian Cancer" (2017). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/6907 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Role of Amylase in Ovarian Cancer by Mai Mohamed A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Pathology and Cell Biology Morsani College of Medicine University of South Florida Major Professor: Patricia Kruk, Ph.D. Paula C. Bickford, Ph.D. Meera Nanjundan, Ph.D. Marzenna Wiranowska, Ph.D. Lauri Wright, Ph.D. Date of Approval: June 29, 2017 Keywords: ovarian cancer, amylase, computational analyses, glycocalyx, cellular invasion Copyright © 2017, Mai Mohamed Dedication This dissertation is dedicated to my parents, Ahmed and Fatma, who have always stressed the importance of education, and, throughout my education, have been my strongest source of encouragement and support. They always believed in me and I am eternally grateful to them. I would also like to thank my brothers, Mohamed and Hussien, and my sister, Mariam. I would also like to thank my husband, Ahmed. -

( 12 ) United States Patent
US010428349B2 (12 ) United States Patent ( 10 ) Patent No. : US 10 , 428 ,349 B2 DeRosa et al . (45 ) Date of Patent: Oct . 1 , 2019 ( 54 ) MULTIMERIC CODING NUCLEIC ACID C12N 2830 / 50 ; C12N 9 / 1018 ; A61K AND USES THEREOF 38 / 1816 ; A61K 38 /45 ; A61K 38/ 44 ; ( 71 ) Applicant: Translate Bio , Inc ., Lexington , MA A61K 38 / 177 ; A61K 48 /005 (US ) See application file for complete search history . (72 ) Inventors : Frank DeRosa , Lexington , MA (US ) ; Michael Heartlein , Lexington , MA (56 ) References Cited (US ) ; Daniel Crawford , Lexington , U . S . PATENT DOCUMENTS MA (US ) ; Shrirang Karve , Lexington , 5 , 705 , 385 A 1 / 1998 Bally et al. MA (US ) 5 ,976 , 567 A 11/ 1999 Wheeler ( 73 ) Assignee : Translate Bio , Inc ., Lexington , MA 5 , 981, 501 A 11/ 1999 Wheeler et al. 6 ,489 ,464 B1 12 /2002 Agrawal et al. (US ) 6 ,534 ,484 B13 / 2003 Wheeler et al. ( * ) Notice : Subject to any disclaimer , the term of this 6 , 815 ,432 B2 11/ 2004 Wheeler et al. patent is extended or adjusted under 35 7 , 422 , 902 B1 9 /2008 Wheeler et al . 7 , 745 ,651 B2 6 / 2010 Heyes et al . U . S . C . 154 ( b ) by 0 days. 7 , 799 , 565 B2 9 / 2010 MacLachlan et al. (21 ) Appl. No. : 16 / 280, 772 7 , 803 , 397 B2 9 / 2010 Heyes et al . 7 , 901, 708 B2 3 / 2011 MacLachlan et al. ( 22 ) Filed : Feb . 20 , 2019 8 , 101 ,741 B2 1 / 2012 MacLachlan et al . 8 , 188 , 263 B2 5 /2012 MacLachlan et al . (65 ) Prior Publication Data 8 , 236 , 943 B2 8 /2012 Lee et al . -

Glycoprotein Hormones and Their Receptors Emerged at the Origin of Metazoans
GBE Glycoprotein Hormones and Their Receptors Emerged at the Origin of Metazoans Graeme J. Roch and Nancy M. Sherwood* Department of Biology, University of Victoria, British Columbia, Canada *Corresponding author: E-mail: [email protected]. Accepted: May 30, 2014 Abstract The cystine knot growth factor (CKGF) superfamily includes important secreted developmental regulators, including the families of transforming growth factor beta, nerve growth factor, platelet-derived growth factor, and the glycoprotein hormones (GPHs). The evolutionary origin of the GPHs and the related invertebrate bursicon hormone, and their characteristic receptors, contributes to an understanding of the endocrine system in metazoans. Using a sensitive search method with hidden Markov models, we identified homologs of the hormones and receptors, along with the closely related bone morphogenetic protein (BMP) antagonists in basal metazoans. In sponges and a comb jelly, cystine knot hormones (CKHs) with mixed features of GPHs, bursicon, and BMP antagonists were identified using primary sequence and phylogenetic analysis. Also, we identified potential receptors for these CKHs, leucine- rich repeat-containing G protein-coupled receptors (LGRs), in the same species. Cnidarians, such as the sea anemone, coral, and hydra, diverged later in metazoan evolution and appear to have duplicated and differentiated CKH-like peptides resulting in bursicon/ GPH-like peptides and several BMP antagonists: Gremlin (Grem), sclerostin domain containing (SOSD), neuroblastoma suppressor of tumorigenicity 1 (NBL1), and Norrie disease protein. An expanded cnidarian LGR group also evolved, including receptors for GPH and bursicon. With the appearance of bilaterians, a separate GPH (thyrostimulin) along with bursicon and BMP antagonists were present. Synteny indicates that the GPHs, Grem, and SOSD have been maintained in a common gene neighborhood throughout much of metazoan evolution. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

Predict AID Targeting in Non-Ig Genes Multiple Transcription Factor
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 is online at: average * The Journal of Immunology published online 20 March 2013 from submission to initial decision 4 weeks from acceptance to publication Multiple Transcription Factor Binding Sites Predict AID Targeting in Non-Ig Genes Jamie L. Duke, Man Liu, Gur Yaari, Ashraf M. Khalil, Mary M. Tomayko, Mark J. Shlomchik, David G. Schatz and Steven H. Kleinstein J Immunol http://www.jimmunol.org/content/early/2013/03/20/jimmun ol.1202547 Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/suppl/2013/03/20/jimmunol.120254 7.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 26, 2021. Published March 20, 2013, doi:10.4049/jimmunol.1202547 The Journal of Immunology Multiple Transcription Factor Binding Sites Predict AID Targeting in Non-Ig Genes Jamie L. Duke,* Man Liu,†,1 Gur Yaari,‡ Ashraf M. Khalil,x Mary M. Tomayko,{ Mark J. Shlomchik,†,x David G. -

Accurate Mutation Annotation and Functional Prediction Enhance the Applicability of -Omics Data in Precision Medicine
The Texas Medical Center Library DigitalCommons@TMC The University of Texas MD Anderson Cancer Center UTHealth Graduate School of The University of Texas MD Anderson Cancer Biomedical Sciences Dissertations and Theses Center UTHealth Graduate School of (Open Access) Biomedical Sciences 5-2016 Accurate mutation annotation and functional prediction enhance the applicability of -omics data in precision medicine Tenghui Chen Follow this and additional works at: https://digitalcommons.library.tmc.edu/utgsbs_dissertations Part of the Bioinformatics Commons, Computational Biology Commons, Genomics Commons, and the Medicine and Health Sciences Commons Recommended Citation Chen, Tenghui, "Accurate mutation annotation and functional prediction enhance the applicability of -omics data in precision medicine" (2016). The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access). 666. https://digitalcommons.library.tmc.edu/utgsbs_dissertations/666 This Dissertation (PhD) is brought to you for free and open access by the The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences at DigitalCommons@TMC. It has been accepted for inclusion in The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access) by an authorized administrator of DigitalCommons@TMC. For more information, please contact [email protected]. ACCURATE MUTATION ANNOTATION AND FUNCTIONAL PREDICTION -
Supplementary Figures S1-S3
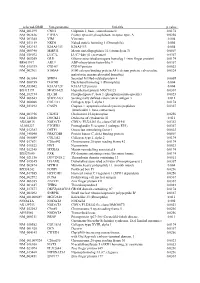
selected-GBID Uni-genename Uni-title p value NM_001299 CNN1 Calponin 1, basic, smooth muscle 0.0174 NM_002836 PTPRA Protein tyrosine phosphatase, receptor type, A 0.0256 NM_003380 VIM Vimentin 0.004 NM_033119 NKD1 Naked cuticle homolog 1 (Drosophila) 0.004 NM_052913 KIAA1913 KIAA1913 0.004 NM_005940 MMP11 Matrix metallopeptidase 11 (stromelysin 3) 0.0069 NM_018032 LUC7L LUC7-like (S. cerevisiae) 0.0367 NM_005269 GLI1 Glioma-associated oncogene homolog 1 (zinc finger protein) 0.0174 BE463997 ARL9 ADP-ribosylation factor-like 9 0.0367 NM_015939 CGI-09 CGI-09 protein 0.0023 NM_002961 S100A4 S100 calcium binding protein A4 (calcium protein, calvasculin, 0.0324 metastasin, murine placental homolog) NM_003014 SFRP4 Secreted frizzled-related protein 4 0.0005 NM_080759 DACH1 Dachshund homolog 1 (Drosophila) 0.004 NM_053042 KIAA1729 KIAA1729 protein 0.004 BX415194 MGC16121 Hypothetical protein MGC16121 0.0367 NM_182734 PLCB1 Phospholipase C, beta 1 (phosphoinositide-specific) 0.0023 NM_006643 SDCCAG3 Serologically defined colon cancer antigen 3 0.011 NM_000088 COL1A1 Collagen, type I, alpha 1 0.0174 NM_033292 CASP1 Caspase 1, apoptosis-related cysteine peptidase 0.0367 (interleukin 1, beta, convertase) NM_003956 CH25H Cholesterol 25-hydroxylase 0.0256 NM_144658 DOCK11 Dedicator of cytokinesis 11 0.011 AK024935 NODATA CDNA: FLJ21283 fis, clone COL01910 0.0363 AL050227 PTGER3 Prostaglandin E receptor 3 (subtype EP3) 0.0367 NM_012383 OSTF1 Osteoclast stimulating factor 1 0.0023 NM_145040 PRKCDBP Protein kinase C, delta binding protein 0.0069 NM_000089 -

Supplementary Figure S1 A
Supplementary Figure S1 A B C 1 4 1 3 1 2 1 2 1 0 1 1 (Normalized signal values)] 2 (Normalized signal values) (Normalized 8 2 10 Log 6 AFFX-BioC AFFX-BioB AFFX-BioDn AFFX-CreX hDFs [Log 6 8 10 12 14 hOFs [Log2 (Normalized signal values)] Supplementary Figure S2 GLYCOLYSIS PENTOSE-PHOSPHATE PATHWAY Glucose Purine/pyrimidine Glucose-6-phosphate metabolism AMINO ACID Fluctose-6-phosphate AMPK METABOLISM TIGAR PFKFB2 methylgloxal GloI Ser, Gly, Thr Glyceraldehyde-3-phosphate ALDH Lactate PYRUVATE LDH METABOLISM acetic acid Ethanol Pyruvate GLYCOSPHINGOLIPID NADH BIOSYNTHESIS Ala, Cys DLD PDH PDK3 DLAT Fatty acid Lys, Trp, Leu, Acetyl CoA ACAT2 Ile, Tyr, Phe β-OXIDATION ACACA Citrate Asp, Asn Citrate Acetyl CoA Oxaloacetate Isocitrate MDH1 IDH1 Glu, Gln, His, ME2 TCA Pro, Arg 2-Oxoglutarate MDH1 CYCLE Pyruvate Malate ME2 GLUTAMINOLYSIS FH Succinyl-CoA Fumalate SUCLA2 Tyr, Phe Var, Ile, Met Supplementary Figure S3 Entrez Gene Symbol Gene Name hODs hDFs hOF-iPSCs GeneID 644 BLVRA biliverdin reductase A 223.9 259.3 253.0 3162 HMOX1 heme oxygenase 1 1474.2 2698.0 452.3 9365 KL klotho 54.1 44.8 36.5 nicotinamide 10135 NAMPT 827.7 626.2 2999.8 phosphoribosyltransferase nuclear factor (erythroid- 4780 NFE2L2 2134.5 1331.7 1006.2 derived 2) related factor 2 peroxisome proliferator- 5467 PPARD 1534.6 1352.9 330.8 activated receptor delta peroxisome proliferator- 5468 PPARG 524.4 100.8 63.0 activated receptor gamma 5621 PRNP prion protein 4059.0 3134.1 1065.5 5925 RB1 retinoblastoma 1 882.9 805.8 739.3 23411 SIRT1 sirtuin 1 231.5 216.8 1676.0 7157 TP53 -

The Role of Gene Regulation in Cancer
Yale University EliScholar – A Digital Platform for Scholarly Publishing at Yale Public Health Theses School of Public Health January 2014 The Role Of Gene Regulation In Cancer: Studies Of Cancer-Related Phenotypes Mediated By Mex3d And By Microrna-618 Implicate Their Potential Oncogenic Role Travis Whitfill Yale University, [email protected] Follow this and additional works at: http://elischolar.library.yale.edu/ysphtdl Recommended Citation Whitfill, Travis, "The Role Of Gene Regulation In Cancer: Studies Of Cancer-Related Phenotypes Mediated By Mex3d And By Microrna-618 Implicate Their otP ential Oncogenic Role" (2014). Public Health Theses. 1322. http://elischolar.library.yale.edu/ysphtdl/1322 This Open Access Thesis is brought to you for free and open access by the School of Public Health at EliScholar – A Digital Platform for Scholarly Publishing at Yale. It has been accepted for inclusion in Public Health Theses by an authorized administrator of EliScholar – A Digital Platform for Scholarly Publishing at Yale. For more information, please contact [email protected]. THE ROLE OF GENE REGULATION IN CANCER: STUDIES OF CANCER- RELATED PHENOTYPES MEDIATED BY MEX3D AND BY MICRORNA-618 IMPLICATE THEIR POTENTIAL ONCOGENIC ROLE A Thesis Presented to the Faculty of the School of Public Health of Yale University in Candidacy for the Degree of Masters of Public Health by Travis Michael Whitfill Primary Thesis Advisor: Yong Zhu May 2014 © 2014 by Travis Michael Whitfill All rights reserved. 2 ABSTRACT The role of gene regulation in cancer: Studies of cancer-related phenotypes mediated by MEX3D and by microRNA-618 implicate their potential oncogenic role Travis Michael Whitfill 2014 The control of gene expression is pivotal in the context of molecular pathogenesis of a number of diseases, and thus is of critical relevance to public health. -

WO 2014/110053 Al 17 July 2014 (17.07.2014) P O P CT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2014/110053 Al 17 July 2014 (17.07.2014) P O P CT (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C12N 7/00 (2006.01) A61K 39/245 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (21) International Application Number: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, PCT/US2014/010553 KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, (22) International Filing Date: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 7 January 2014 (07.01 .2014) OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (25) Filing Language: English TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, (26) Publication Language: English ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 61/750,175 8 January 20 13 (08.01 .2013) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: GENZYME CORPORATION [US/US]; 500 UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, Kendall Square, Cambridge, MA 02142 (US). TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, (72) Inventors; and MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, (71) Applicants : PECHAN, Peter [US/US]; 500 Kendall TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Square, Cambridge, MA 02141 (US). -

Nonadditive Effects of Genes in Human Metabolomics
HIGHLIGHTED ARTICLE GENETICS | INVESTIGATION Nonadditive Effects of Genes in Human Metabolomics Yakov A. Tsepilov,*,†,‡,§,** So-Youn Shin,††,‡‡ Nicole Soranzo,††,§§ Tim D. Spector,*** Cornelia Prehn,††† Jerzy Adamski,†††,‡‡‡,§§§ Gabi Kastenmüller,***,**** Rui Wang-Sattler,§,** Konstantin Strauch,‡,†††† Christian Gieger,‡,**,§ Yurii S. Aulchenko,*,† and Janina S. Ried‡ *Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences, 630090 Novosibirsk, Russia, †Novosibirsk State University, 630090 Novosibirsk, Russia, ‡Institute of Genetic Epidemiology, §Research Unit of Molecular Epidemiology, **Institute of Epidemiology II, †††Institute of Experimental Genetics, Genome Analysis Center, and ****Institute of Bioinformatics and Systems Biology, Helmholtz Zentrum München–German Research Center for Environmental Health, 85764 Neuherberg, Germany, ††Human Genetics, Wellcome Trust Sanger Institute, Genome Campus, Hinxton, CB10 1HH, United Kingdom, ‡‡MRC Integrative Epidemiology Unit, School of Social and Community Medicine, University of Bristol, Bristol, BS8 1TH, United Kingdom, §§Department of Haematology, University of Cambridge, Cambridge, CB2 0AH, United Kingdom, ***Department of Twin Research and Genetic Epidemiology, King’s College London, London, WC2R 2LS, United Kingdom, ‡‡‡Institute of Experimental Genetics, Life and Food Science Center Weihenstephan, Technische Universität München, 85354 Freising-Weihenstephan, Germany, §§§German Center for Diabetes Research, 85764 Neuherberg, Germany, ††††Institute -

Whole-Exome Sequencing Validates a Preclinical Mouse Model for the Prevention and Treatment of Cutaneous Squamous Cell Carcinoma Elena V
Published OnlineFirst December 6, 2016; DOI: 10.1158/1940-6207.CAPR-16-0218 Research Article Cancer Prevention Research Whole-Exome Sequencing Validates a Preclinical Mouse Model for the Prevention and Treatment of Cutaneous Squamous Cell Carcinoma Elena V. Knatko1, Brandon Praslicka2, Maureen Higgins1, Alan Evans3, Karin J. Purdie4, Catherine A. Harwood4, Charlotte M. Proby1, Aikseng Ooi2, and Albena T. Dinkova-Kostova1,5,6 Abstract Cutaneous squamous cell carcinomas (cSCC) are among the detected in 15 of 18 (83%) cases, with 20 of 21 SNP mutations most common and highly mutated human malignancies. Solar located in the protein DNA-binding domain. Strikingly, multiple UV radiation is the major factor in the etiology of cSCC. Whole- nonsynonymous SNP mutations in genes encoding Notch family exome sequencing of 18 microdissected tumor samples (cases) members (Notch1-4) were present in 10 of 18 (55%) cases. The derived from SKH-1 hairless mice that had been chronically histopathologic spectrum of the mouse cSCC that develops in this exposed to solar-simulated UV (SSUV) radiation showed a medi- model resembles very closely the spectrum of human cSCC. We an point mutation (SNP) rate of 155 per Mb. The majority conclude that the mouse SSUV cSCCs accurately represent the (78.6%) of the SNPs are C.G>T.A transitions, a characteristic histopathologic and mutational spectra of the most prevalent UVR-induced mutational signature. Direct comparison with tumor suppressors of human cSCC, validating the use of this human cSCC cases showed high overlap in terms of both fre- preclinical model for the prevention and treatment of human quency and type of SNP mutations.