Open Science in Machine Learning
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CNRS ROADMAP for OPEN SCIENCE 18 November 2019
CNRS ROADMAP FOR OPEN SCIENCE 18 November 2019 TABLE OF CONTENTS Introduction 4 1. Publications 6 2. Research data 8 3. Text and data mining and analysis 10 4. Individual evaluation of researchers and Open Science 11 5. Recasting Scientific and Technical Information for Open Science 12 6. Training and skills 13 7. International positioning 14 INTRODUCTION The international movement towards Open Science started more than 30 years ago and has undergone unprecedented development since the web made it possible on a global scale with reasonable costs. The dissemination of scientific production on the Internet, its identification and archiving lift the barriers to permanent access without challenging the protection of personal data or intellectual property. Now is the time to make it “as open as possible, as closed as necessary”. Open Science is not only about promoting a transversal approach to the sharing of scientific results. By opening up data, processes, codes, methods or protocols, it also offers a new way of doing science. Several scientific, civic and socio-economic reasons make Just over a year ago, France embarked on this vast transfor- the development of Open Science essential today: mation movement. Presented on 4 July 2018 by the Minister • Sharing scientific knowledge makes research more ef- of Higher Education,Research and Innovation, the “Natio- fective, more visible, and less redundant. Open access to nal Plan for Open Science”1 aims, in the words of Frédérique data and results is a sea change for the way research is Vidal, to ensure that “the results of scientific research are done, and opens the way to the use of new tools. -

Understanding the Value of Arts & Culture | the AHRC Cultural Value
Understanding the value of arts & culture The AHRC Cultural Value Project Geoffrey Crossick & Patrycja Kaszynska 2 Understanding the value of arts & culture The AHRC Cultural Value Project Geoffrey Crossick & Patrycja Kaszynska THE AHRC CULTURAL VALUE PROJECT CONTENTS Foreword 3 4. The engaged citizen: civic agency 58 & civic engagement Executive summary 6 Preconditions for political engagement 59 Civic space and civic engagement: three case studies 61 Part 1 Introduction Creative challenge: cultural industries, digging 63 and climate change 1. Rethinking the terms of the cultural 12 Culture, conflict and post-conflict: 66 value debate a double-edged sword? The Cultural Value Project 12 Culture and art: a brief intellectual history 14 5. Communities, Regeneration and Space 71 Cultural policy and the many lives of cultural value 16 Place, identity and public art 71 Beyond dichotomies: the view from 19 Urban regeneration 74 Cultural Value Project awards Creative places, creative quarters 77 Prioritising experience and methodological diversity 21 Community arts 81 Coda: arts, culture and rural communities 83 2. Cross-cutting themes 25 Modes of cultural engagement 25 6. Economy: impact, innovation and ecology 86 Arts and culture in an unequal society 29 The economic benefits of what? 87 Digital transformations 34 Ways of counting 89 Wellbeing and capabilities 37 Agglomeration and attractiveness 91 The innovation economy 92 Part 2 Components of Cultural Value Ecologies of culture 95 3. The reflective individual 42 7. Health, ageing and wellbeing 100 Cultural engagement and the self 43 Therapeutic, clinical and environmental 101 Case study: arts, culture and the criminal 47 interventions justice system Community-based arts and health 104 Cultural engagement and the other 49 Longer-term health benefits and subjective 106 Case study: professional and informal carers 51 wellbeing Culture and international influence 54 Ageing and dementia 108 Two cultures? 110 8. -

Open Educational Practices and Resources
Open Educational Practices and Resources OLCOS Roadmap 2012 Edited by Guntram Geser Salzburg Research EduMedia Group Project information and imprint Project information and imprint Open e-Learning Content Observatory Services (OLCOS) OLCOS is a Transversal Action funded by the European Commission under the eLearning Programme. Duration: January 2006 – December 2007 Website: www.olcos.org Project partners European Centre for Media Competence, Germany European Distance and E-Learning Network, Hungary FernUniversitaet in Hagen, Germany Mediamaisteri Group, Finland Open University of Catalonia, Spain Salzburg Research, Austria Project coordinator Salzburg Research / EduMedia Group Veronika Hornung-Prähauser Jakob Haringer Straße 5/III, A-5020 Salzburg, Austria [email protected] Tel. 0043-662-2288-405 OLCOS roadmap editor Guntram Geser, Salzburg Research / EduMedia Group, Austria Contributors to the OLCOS roadmap FernUniversitaet in Hagen: Peter Baumgartner and Viola Naust Open University of Catalonia: Agustí Canals, Núria Ferran, Julià Minguillón and Mireia Pascual Mediamaisteri Group: Mats Rajalakso and Timo Väliharju Salzburg Research: Wernher Behrendt, Andreas Gruber, Veronika Hornung-Prähauser and Sebastian Schaffert Graphics & layout Jesper Visser, Salzburg Research 3 Project information and imprint Images Based on copyright-free photographs from www.imageafter.com Print version ISBN 3-902448-08-3 Printed in Austria January 2007 Online A digital version of this report can be freely downloaded from www.olcos.org Copyright This work is licensed under the Creative Commons Attribution–NonCommercial–ShareAlike 2.5 License http://creativecommons.org/licenses/by-nc-sa/2.5/ Disclaimer This publication was produced by the OLCOS Project with the financial support of the European Commission. The content of this report is the sole responsibility of OLCOS and its project partners. -

Is Sci-Hub Increasing Visibility of Indian Research Papers? an Analytical Evaluation Vivek Kumar Singh1,*, Satya Swarup Srichandan1, Sujit Bhattacharya2
Journal of Scientometric Res. 2021; 10(1):130-134 http://www.jscires.org Perspective Paper Is Sci-Hub Increasing Visibility of Indian Research Papers? An Analytical Evaluation Vivek Kumar Singh1,*, Satya Swarup Srichandan1, Sujit Bhattacharya2 1Department of Computer Science, Banaras Hindu University, Varanasi, Uttar Pradesh, INDIA. 2CSIR-National Institute of Science Technology and Development Studies, New Delhi, INDIA. ABSTRACT Sci-Hub, founded by Alexandra Elbakyan in 2011 in Kazakhstan has, over the years, Correspondence emerged as a very popular source for researchers to download scientific papers. It is Vivek Kumar Singh believed that Sci-Hub contains more than 76 million academic articles. However, recently Department of Computer Science, three foreign academic publishers (Elsevier, Wiley and American Chemical Society) have Banaras Hindu University, filed a lawsuit against Sci-Hub and LibGen before the Delhi High Court and prayed for Varanasi-221005, INDIA. complete blocking these websites in India. It is in this context, that this paper attempts to Email id: [email protected] find out how many Indian research papers are available in Sci-Hub and who downloads them. The citation advantage of Indian research papers available on Sci-Hub is analysed, Received: 16-03-2021 with results confirming that such an advantage do exist. Revised: 29-03-2021 Accepted: 25-04-2021 Keywords: Indian Research, Indian Science, Black Open Access, Open Access, Sci-Hub. DOI: 10.5530/jscires.10.1.16 INTRODUCTION access publishing of their research output, and at the same time encouraging their researchers to publish in openly Responsible Research and Innovation (RRI) has become one accessible forms. -

A Comprehensive Framework to Reinforce Evidence Synthesis Features in Cloud-Based Systematic Review Tools
applied sciences Article A Comprehensive Framework to Reinforce Evidence Synthesis Features in Cloud-Based Systematic Review Tools Tatiana Person 1,* , Iván Ruiz-Rube 1 , José Miguel Mota 1 , Manuel Jesús Cobo 1 , Alexey Tselykh 2 and Juan Manuel Dodero 1 1 Department of Informatics Engineering, University of Cadiz, 11519 Puerto Real, Spain; [email protected] (I.R.-R.); [email protected] (J.M.M.); [email protected] (M.J.C.); [email protected] (J.M.D.) 2 Department of Information and Analytical Security Systems, Institute of Computer Technologies and Information Security, Southern Federal University, 347922 Taganrog, Russia; [email protected] * Correspondence: [email protected] Abstract: Systematic reviews are powerful methods used to determine the state-of-the-art in a given field from existing studies and literature. They are critical but time-consuming in research and decision making for various disciplines. When conducting a review, a large volume of data is usually generated from relevant studies. Computer-based tools are often used to manage such data and to support the systematic review process. This paper describes a comprehensive analysis to gather the required features of a systematic review tool, in order to support the complete evidence synthesis process. We propose a framework, elaborated by consulting experts in different knowledge areas, to evaluate significant features and thus reinforce existing tool capabilities. The framework will be used to enhance the currently available functionality of CloudSERA, a cloud-based systematic review Citation: Person, T.; Ruiz-Rube, I.; Mota, J.M.; Cobo, M.J.; Tselykh, A.; tool focused on Computer Science, to implement evidence-based systematic review processes in Dodero, J.M. -
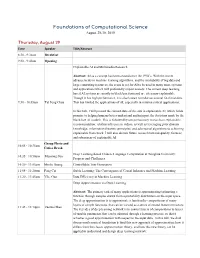
Foundations of Computational Science August 29-30, 2019
Foundations of Computational Science August 29-30, 2019 Thursday, August 29 Time Speaker Title/Abstract 8:30 - 9:30am Breakfast 9:30 - 9:40am Opening Explainable AI and Multimedia Research Abstract: AI as a concept has been around since the 1950’s. With the recent advancements in machine learning algorithms, and the availability of big data and large computing resources, the scene is set for AI to be used in many more systems and applications which will profoundly impact society. The current deep learning based AI systems are mostly in black box form and are often non-explainable. Though it has high performance, it is also known to make occasional fatal mistakes. 9:30 - 10:05am Tat Seng Chua This has limited the applications of AI, especially in mission critical applications. In this talk, I will present the current state-of-the arts in explainable AI, which holds promise to helping humans better understand and interpret the decisions made by the black-box AI models. This is followed by our preliminary research on explainable recommendation, relation inference in videos, as well as leveraging prior domain knowledge, information theoretic principles, and adversarial algorithms to achieving explainable framework. I will also discuss future research towards quality, fairness and robustness of explainable AI. Group Photo and 10:05 - 10:35am Coffee Break Deep Learning-based Chinese Language Computation at Tsinghua University: 10:35 - 10:50am Maosong Sun Progress and Challenges 10:50 - 11:05am Minlie Huang Controllable Text Generation 11:05 - 11:20am Peng Cui Stable Learning: The Convergence of Causal Inference and Machine Learning 11:20 - 11:45am Yike Guo Data Efficiency in Machine Learning Deep Approximation via Deep Learning Abstract: The primary task of many applications is approximating/estimating a function through samples drawn from a probability distribution on the input space. -

From Coalition to Commons: Plan S and the Future of Scholarly Communication
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Copyright, Fair Use, Scholarly Communication, etc. Libraries at University of Nebraska-Lincoln 2019 From Coalition to Commons: Plan S and the Future of Scholarly Communication Rob Johnson Research Consulting Follow this and additional works at: https://digitalcommons.unl.edu/scholcom Part of the Intellectual Property Law Commons, Scholarly Communication Commons, and the Scholarly Publishing Commons Johnson, Rob, "From Coalition to Commons: Plan S and the Future of Scholarly Communication" (2019). Copyright, Fair Use, Scholarly Communication, etc.. 157. https://digitalcommons.unl.edu/scholcom/157 This Article is brought to you for free and open access by the Libraries at University of Nebraska-Lincoln at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Copyright, Fair Use, Scholarly Communication, etc. by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Insights – 32, 2019 Plan S and the future of scholarly communication | Rob Johnson From coalition to commons: Plan S and the future of scholarly communication The announcement of Plan S in September 2018 triggered a wide-ranging debate over how best to accelerate the shift to open access. The Plan’s ten principles represent a call for the creation of an intellectual commons, to be brought into being through collective action by funders and managed through regulated market mechanisms. As it gathers both momentum and critics, the coalition must grapple with questions of equity, efficiency and sustainability. The work of Elinor Ostrom has shown that successful management of the commons frequently relies on polycentricity and adaptive governance. The Plan S principles must therefore function as an overarching framework within which local actors retain some autonomy, and should remain open to amendment as the scholarly communication landscape evolves. -

Open Access Availability of Scientific Publications
Analytical Support for Bibliometrics Indicators Open access availability of scientific publications Analytical Support for Bibliometrics Indicators Open access availability of scientific publications* Final Report January 2018 By: Science-Metrix Inc. 1335 Mont-Royal E. ▪ Montréal ▪ Québec ▪ Canada ▪ H2J 1Y6 1.514.495.6505 ▪ 1.800.994.4761 [email protected] ▪ www.science-metrix.com *This work was funded by the National Science Foundation’s (NSF) National Center for Science and Engineering Statistics (NCSES). Any opinions, findings, conclusions or recommendations expressed in this report do not necessarily reflect the views of NCSES or the NSF. The analysis for this research was conducted by SRI International on behalf of NSF’s NCSES under contract number NSFDACS1063289. Analytical Support for Bibliometrics Indicators Open access availability of scientific publications Contents Contents .............................................................................................................................................................. i Tables ................................................................................................................................................................. ii Figures ................................................................................................................................................................ ii Abstract ............................................................................................................................................................ -

Radical Solutions and Open Science an Open Approach to Boost Higher Education Lecture Notes in Educational Technology
Lecture Notes in Educational Technology Daniel Burgos Editor Radical Solutions and Open Science An Open Approach to Boost Higher Education Lecture Notes in Educational Technology Series Editors Ronghuai Huang, Smart Learning Institute, Beijing Normal University, Beijing, China Kinshuk, College of Information, University of North Texas, Denton, TX, USA Mohamed Jemni, University of Tunis, Tunis, Tunisia Nian-Shing Chen, National Yunlin University of Science and Technology, Douliu, Taiwan J. Michael Spector, University of North Texas, Denton, TX, USA The series Lecture Notes in Educational Technology (LNET), has established itself as a medium for the publication of new developments in the research and practice of educational policy, pedagogy, learning science, learning environment, learning resources etc. in information and knowledge age, – quickly, informally, and at a high level. Abstracted/Indexed in: Scopus, Web of Science Book Citation Index More information about this series at http://www.springer.com/series/11777 Daniel Burgos Editor Radical Solutions and Open Science An Open Approach to Boost Higher Education Editor Daniel Burgos Research Institute for Innovation & Technology in Education (UNIR iTED) Universidad Internacional de La Rioja (UNIR) Logroño, La Rioja, Spain ISSN 2196-4963 ISSN 2196-4971 (electronic) Lecture Notes in Educational Technology ISBN 978-981-15-4275-6 ISBN 978-981-15-4276-3 (eBook) https://doi.org/10.1007/978-981-15-4276-3 © The Editor(s) (if applicable) and The Author(s) 2020. This book is an open access publication. Open Access This book is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made. -

Computational Science and Engineering
Computational Science and Engineering, PhD College of Engineering Graduate Coordinator: TBA Email: Phone: Department Chair: Marwan Bikdash Email: [email protected] Phone: 336-334-7437 The PhD in Computational Science and Engineering (CSE) is an interdisciplinary graduate program designed for students who seek to use advanced computational methods to solve large problems in diverse fields ranging from the basic sciences (physics, chemistry, mathematics, etc.) to sociology, biology, engineering, and economics. The mission of Computational Science and Engineering is to graduate professionals who (a) have expertise in developing novel computational methodologies and products, and/or (b) have extended their expertise in specific disciplines (in science, technology, engineering, and socioeconomics) with computational tools. The Ph.D. program is designed for students with graduate and undergraduate degrees in a variety of fields including engineering, chemistry, physics, mathematics, computer science, and economics who will be trained to develop problem-solving methodologies and computational tools for solving challenging problems. Research in Computational Science and Engineering includes: computational system theory, big data and computational statistics, high-performance computing and scientific visualization, multi-scale and multi-physics modeling, computational solid, fluid and nonlinear dynamics, computational geometry, fast and scalable algorithms, computational civil engineering, bioinformatics and computational biology, and computational physics. Additional Admission Requirements Master of Science or Engineering degree in Computational Science and Engineering (CSE) or in science, engineering, business, economics, technology or in a field allied to computational science or computational engineering field. GRE scores Program Outcomes: Students will demonstrate critical thinking and ability in conducting research in engineering, science and mathematics through computational modeling and simulations. -

Artificial Intelligence Research For
Charles Xie ([email protected]) is a senior scientist who works at the inter- section between computational science and learning science. Artificial Intelligence research for engineering design By Charles Xie Imagine a high school classroom in 2030. AI is one of the three most promising areas for which Bill Gates Students are challenged to design an said earlier this year he would drop out of college again if he were a young student today. The victory of Google’s AlphaGo against engineering system that meets the needs of a top human Go player in 2016 was a landmark in the history of their community. They work with advanced AI. The game of Go is considered a difficult challenge because computer-aided design (CAD) software that the number of legal positions on a 19×19 grid is estimated to be leverages the power of scientific simulation, 2×10170, far more than the total number of atoms in the observ- mixed reality, and cloud computing. Every able universe combined (1082). While AlphaGo may be only a cool tool needed to complete an engineering type of weak AI that plays Go at present, scientists believe that its development will engender technologies that eventually find design project is seamlessly integrated into applications in other fields. the CAD platform to streamline the learning process. But the most striking capability of Performance modeling the software is its ability to act like a mentor (e.g., simulation-based analysis) who has all the knowledge about the design project to answer questions, learns from all the successful or unsuccessful solutions ever Evaluator attempted by human designers or computer agents, adapts to the needs of each student based on experience interacting with all sorts of designers, inspires students to explore creative ideas, and, most importantly, remains User Ideator responsive all the time without ever running out of steam. -

Technical Writing
Technical Writing Engineers and scientists perform many complex and intricate tasks using the world's most sophisticated equipment. However, their performance as engineers and scientists is almost always related to their use of one of the oldest tools - the pen. In academia, the saying "publish or perish" often describes the process of acquiring tenure as well as credibility. In industry, both large and small organizations communicate everything through memos, reports, and short presentations. Product development decisions are often made by a committee of people far removed from the actual technology. The saying "he who has the most convincing viewgraphs and reports, wins..." can sometimes apply to industry. Therefore, it should be clear that an ability to concisely and efficiently prepare technical reports, research papers, and or viewgraph presentations can have a profound positive impact on an individual's career. Consider the following statement by anonymous Fortune 500 corporate vice president: "... in any large organization, the person who decides whether you get a promotion, or who determines the size of a pay raise, does not know you personally. The only thing they have to go on is what other people write about you and what you write about you ..." It can be seen that if one should write a lot of material to get ahead in one's career, it makes sense to write as objectively and concisely as possible. Objective writing is essential because good technical writing should not be seen as erroneous after new discoveries are made. A good technical report should present a clear milestone of what was done and understood at the time of the writing.