Immunological Properties and Cdna Sequence Analysis of an Intermediate-filament-Like Protein from Squid Neuronal Tissue
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Blood Neurofilament Light Chain: the Neurologist's Troponin?
biomedicines Review Blood Neurofilament Light Chain: The Neurologist’s Troponin? Simon Thebault 1,*, Ronald A. Booth 2 and Mark S. Freedman 1,* 1 Department of Medicine and the Ottawa Hospital Research Institute, The University of Ottawa, Ottawa, ON K1H8L6, Canada 2 Department of Pathology and Laboratory Medicine, Eastern Ontario Regional Laboratory Association and Ottawa Hospital Research Institute, University of Ottawa & The Ottawa Hospital, Ottawa, ON K1H8L6, Canada; [email protected] * Correspondence: [email protected] (S.T.); [email protected] (M.S.F.) Received: 4 November 2020; Accepted: 18 November 2020; Published: 21 November 2020 Abstract: Blood neurofilament light chain (NfL) is a marker of neuro-axonal injury showing promising associations with outcomes of interest in several neurological conditions. Although initially discovered and investigated in the cerebrospinal fluid (CSF), the recent development of ultrasensitive digital immunoassay technologies has enabled reliable detection in serum/plasma, obviating the need for invasive lumbar punctures for longitudinal assessment. The most evidence for utility relates to multiple sclerosis (MS) where it serves as an objective measure of both the inflammatory and degenerative pathologies that characterise this disease. In this review, we summarise the physiology and pathophysiology of neurofilaments before focusing on the technological advancements that have enabled reliable quantification of NfL in blood. As the test case for clinical translation, we then highlight important recent developments linking blood NfL levels to outcomes in MS and the next steps to be overcome before this test is adopted on a routine clinical basis. Keywords: neurofilament light chain; biomarkers; multiple sclerosis 1. Neurofilament Structure and Function Neurofilaments are neuronal-specific heteropolymers conventionally considered to consist of a triplet of light (NfL), medium (NfM) and heavy (NfH) chains according to their molecular mass [1]. -

Neurofilaments: Neurobiological Foundations for Biomarker Applications
Neurofilaments: neurobiological foundations for biomarker applications Arie R. Gafson1, Nicolas R. Barthelmy2*, Pascale Bomont3*, Roxana O. Carare4*, Heather D. Durham5*, Jean-Pierre Julien6,7*, Jens Kuhle8*, David Leppert8*, Ralph A. Nixon9,10,11,12*, Roy Weller4*, Henrik Zetterberg13,14,15,16*, Paul M. Matthews1,17 1 Department of Brain Sciences, Imperial College, London, UK 2 Department of Neurology, Washington University School of Medicine, St Louis, MO, USA 3 a ATIP-Avenir team, INM, INSERM , Montpellier university , Montpellier , France. 4 Clinical Neurosciences, Faculty of Medicine, University of Southampton, Southampton General Hospital, Southampton, United Kingdom 5 Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal, Québec, Canada 6 Department of Psychiatry and Neuroscience, Laval University, Quebec, Canada. 7 CERVO Brain Research Center, 2601 Chemin de la Canardière, Québec, QC, G1J 2G3, Canada 8 Neurologic Clinic and Policlinic, Departments of Medicine, Biomedicine and Clinical Research, University Hospital Basel, University of Basel, Basel, Switzerland. 9 Center for Dementia Research, Nathan Kline Institute, Orangeburg, NY, 10962, USA. 10Departments of Psychiatry, New York University School of Medicine, New York, NY, 10016, 11 Neuroscience Institute, New York University School of Medicine, New York, NY, 10016, USA. 12Department of Cell Biology, New York University School of Medicine, New York, NY, 10016, USA 13 University College London Queen Square Institute of Neurology, London, UK 14 UK Dementia Research Institute at University College London 15 Department of Psychiatry and Neurochemistry, Institute of Neuroscience and Physiology, the Sahlgrenska Academy at the University of Gothenburg, Mölndal, Sweden 16 Clinical Neurochemistry Laboratory, Sahlgrenska University Hospital, Mölndal, Sweden 17 UK Dementia Research Institute at Imperial College, London * Co-authors ordered alphabetically Address for correspondence: Prof. -

Intermediate Filament Accumulation Can Stabilize Microtubules in Caenorhabditis Elegans Motor Neurons
Intermediate filament accumulation can stabilize microtubules in Caenorhabditis elegans motor neurons Naina Kurupa, Yunbo Lia, Alexandr Goncharova, and Yishi Jina,b,1 aNeurobiology Section, Division of Biological Sciences, University of California, San Diego, La Jolla, CA 92093; and bDepartment of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093 Edited by H. Robert Horvitz, Massachusetts Institute of Technology, Cambridge, MA, and approved February 11, 2018 (received for review December 21, 2017) Neural circuits utilize a coordinated cellular machinery to form and Results eliminate synaptic connections, with the neuronal cytoskeleton Identification of IF Genes That Regulate Synapse Rewiring. At the playing a prominent role. During larval development of Caenorhabditis end of larval stage 1 (L1), the dorsal D (DD)-type motor neurons elegans, synapses of motor neurons are stereotypically rewired rewire their presynaptic connections from the ventral nerve cord through a process facilitated by dynamic microtubules (MTs). Through a (VNC) to the dorsal nerve cord (DNC), concurrent with the genetic suppressor screen on mutant animals that fail to rewire synap- birth of ventral D (VD)-type motor neurons, which then form ses, and in combination with live imaging and ultrastructural studies, synapses along the VNC (19). We visualized DD-neuron pre- we find that intermediate filaments (IFs) stabilize MTs to prevent syn- synaptic terminals using a GFP-tagged synaptobrevin (SNB- apse rewiring. Genetic ablation of IFs or pharmacological disruption of 1::GFP) reporter (juIs137:Pflp-13 SNB-1::GFP). In L1 animals, IF networks restores MT growth and rescues synapse rewiring defects discrete synaptic puncta were present along the ventral neurites in the mutant animals, indicating that IF accumulation directly alters MT (18), but in late larvae and adults, synaptic puncta were only seen stability. -

Attenuated Neurodegenerative Disease Phenotype in Tau Transgenic Mouse Lacking Neurofilaments
The Journal of Neuroscience, August 15, 2001, 21(16):6026–6035 Attenuated Neurodegenerative Disease Phenotype in Tau Transgenic Mouse Lacking Neurofilaments Takeshi Ishihara, Makoto Higuchi, Bin Zhang, Yasumasa Yoshiyama, Ming Hong, John Q. Trojanowski, and Virginia M.-Y. Lee Center for Neurodegenerative Disease Research, Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104 Previous studies have shown that transgenic (Tg) mice overex- particular NFL, resulted in a dramatic decrease in the total pressing human tau protein develop filamentous tau aggre- number of tau-positive spheroids in spinal cord and brainstem. gates in the CNS. The most abundant tau aggregates are found Concomitant with the reduction in spheroid number, the bigenic in spinal cord and brainstem in which they colocalize with mice showed delayed accumulation of insoluble tau protein in neurofilaments (NFs) as spheroids in axons. To elucidate the the CNS, increased viability, reduced weight loss, and improved role of NF subunit proteins in tau aggregate formation and to behavioral phenotype when compared with the single T44 Tg test the hypothesis that NFs are pathological chaperones in the mice. These results imply that NFs are pathological chaperones formation of intraneuronal tau inclusions, we crossbred previ- in the development of tau spheroids and suggest a role for NFs ously described tau (T44) Tg mice overexpressing the smallest in the pathogenesis of neurofibrillary tau lesions in neurodegen- -

Neurofilaments and Neurofilament Proteins in Health and Disease
Downloaded from http://cshperspectives.cshlp.org/ on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press Neurofilaments and Neurofilament Proteins in Health and Disease Aidong Yuan,1,2 Mala V. Rao,1,2 Veeranna,1,2 and Ralph A. Nixon1,2,3 1Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York 10962 2Department of Psychiatry, New York University School of Medicine, New York, New York 10016 3Cell Biology, New York University School of Medicine, New York, New York 10016 Correspondence: [email protected], [email protected] SUMMARY Neurofilaments (NFs) are unique among tissue-specific classes of intermediate filaments (IFs) in being heteropolymers composed of four subunits (NF-L [neurofilament light]; NF-M [neuro- filament middle]; NF-H [neurofilament heavy]; and a-internexin or peripherin), each having different domain structures and functions. Here, we review how NFs provide structural support for the highly asymmetric geometries of neurons and, especially, for the marked radial expan- sion of myelinated axons crucial for effective nerve conduction velocity. NFs in axons exten- sively cross-bridge and interconnect with other non-IF components of the cytoskeleton, including microtubules, actin filaments, and other fibrous cytoskeletal elements, to establish a regionallyspecialized networkthat undergoes exceptionallyslow local turnoverand serves as a docking platform to organize other organelles and proteins. We also discuss how a small pool of oligomeric and short filamentous precursors in the slow phase of axonal transport maintains this network. A complex pattern of phosphorylation and dephosphorylation events on each subunit modulates filament assembly, turnover, and organization within the axonal cytoskel- eton. Multiple factors, and especially turnover rate, determine the size of the network, which can vary substantially along the axon. -

The Role of Tau in Amyloid Β Plaque Growth and Formation
THE ROLE OF TAU IN AMYLOID β PLAQUE GROWTH AND FORMATION Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades an der Fakultät für Biologie der Ludwig-Maximilians-Universität München vorgelegt von Hazal Salihoglu München, den 07.06.2019 i Neyi arıyorsan O’sun sen. What you seek is seeking you. Was du suchst, sucht dich. Mevlana (1207-1273) i Diese Dissertation wurde unter der Leitung von Prof. Dr. Jochen Herms am Institut für Neuropathologie der Ludwig-Maximilians-Universität angefertigt und von Prof. Dr. Wolfgang Enard an der Fakultät für Biologie vertreten. Eidesstattliche Erklärung Ich versichere hiermit an Eides statt, dass die vorgelegte Dissertation von mir selbständig und ohne unerlaubte Hilfe angefertigt ist. Hiermit erkläre ich, dass die Dissertation nicht ganz oder in wesentlichen Teilen einer anderen Prüfungskommission vorgelegt worden ist und ich mich anderweitig einer Doktorprüfung ohne Erfolg nicht unterzogen habe. München, den 17.10.2018 Hazal Salihoglu Erstgutachter: Prof. Dr. Wolfgang Enard Zweitgutachter: Dr. rer. nat. Anja Horn-Bochtler Tag der Abgabe: 06.11.2018 Tag der mündlichen Prüfung: 27.05.19 ii Table of contents Table of Contents ACKNOWLEDGEMENTS .................................................................................................... 1 DECLARATION ................................................................................................................... 2 LIST OF FIGURES .............................................................................................................. -

Cytoskeletal Proteins in Neurological Disorders
cells Review Much More Than a Scaffold: Cytoskeletal Proteins in Neurological Disorders Diana C. Muñoz-Lasso 1 , Carlos Romá-Mateo 2,3,4, Federico V. Pallardó 2,3,4 and Pilar Gonzalez-Cabo 2,3,4,* 1 Department of Oncogenomics, Academic Medical Center, 1105 AZ Amsterdam, The Netherlands; [email protected] 2 Department of Physiology, Faculty of Medicine and Dentistry. University of Valencia-INCLIVA, 46010 Valencia, Spain; [email protected] (C.R.-M.); [email protected] (F.V.P.) 3 CIBER de Enfermedades Raras (CIBERER), 46010 Valencia, Spain 4 Associated Unit for Rare Diseases INCLIVA-CIPF, 46010 Valencia, Spain * Correspondence: [email protected]; Tel.: +34-963-395-036 Received: 10 December 2019; Accepted: 29 January 2020; Published: 4 February 2020 Abstract: Recent observations related to the structure of the cytoskeleton in neurons and novel cytoskeletal abnormalities involved in the pathophysiology of some neurological diseases are changing our view on the function of the cytoskeletal proteins in the nervous system. These efforts allow a better understanding of the molecular mechanisms underlying neurological diseases and allow us to see beyond our current knowledge for the development of new treatments. The neuronal cytoskeleton can be described as an organelle formed by the three-dimensional lattice of the three main families of filaments: actin filaments, microtubules, and neurofilaments. This organelle organizes well-defined structures within neurons (cell bodies and axons), which allow their proper development and function through life. Here, we will provide an overview of both the basic and novel concepts related to those cytoskeletal proteins, which are emerging as potential targets in the study of the pathophysiological mechanisms underlying neurological disorders. -

Specialized Roles of Neurofilament Proteins in Synapses
Brain Research Bulletin 126 (2016) 334–346 Contents lists available at ScienceDirect Brain Research Bulletin j ournal homepage: www.elsevier.com/locate/brainresbull Review Specialized roles of neurofilament proteins in synapses: Relevance to neuropsychiatric disorders a,b,∗ a,b,c,∗ Aidong Yuan , Ralph A. Nixon a Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, 10962, United States b Departments of Psychiatry, New York University School of Medicine, New York, NY, 10016, United States c Department of Cell Biology, New York University School of Medicine, New York, NY, 10016, United States a r t i c l e i n f o a b s t r a c t Article history: Neurofilaments are uniquely complex among classes of intermediate filaments in being composed of four Received 11 July 2016 subunits (NFL, NFM, NFH and alpha-internexin in the CNS) that differ in structure, regulation, and func- Received in revised form 2 September 2016 tion. Although neurofilaments have been traditionally viewed as axonal structural components, recent Accepted 3 September 2016 evidence has revealed that distinctive assemblies of neurofilament subunits are integral components Available online 5 September 2016 of synapses, especially at postsynaptic sites. Within the synaptic compartment, the individual subunits differentially modulate neurotransmission and behavior through interactions with specific neurotrans- Keywords: mitter receptors. These newly uncovered functions suggest that alterations of neurofilament proteins Neurofilament subunit Synapse not only underlie axonopathy in various neurological disorders but also may play vital roles in cog- nition and neuropsychiatric diseases. Here, we review evidence that synaptic neurofilament proteins Dendritic spine Neuropsychiatric disease are a sizable population in the CNS and we advance the concept that changes in the levels or post- translational modification of individual NF subunits contribute to synaptic and behavioral dysfunction in certain neuropsychiatric conditions. -

Internexin Causes Abnormal Neurofilamentous Accumulations
The Journal of Neuroscience, April 15, 1999, 19(8):2974–2986 Overexpression of a-Internexin Causes Abnormal Neurofilamentous Accumulations and Motor Coordination Deficits in Transgenic Mice Gee Y. Ching,1 Chung-Liang Chien,2 Roberto Flores,1 and Ronald K. H. Liem1 1Departments of Pathology and Anatomy and Cell Biology, Columbia University College of Physicians and Surgeons, New York, New York 10032, and 2Department of Anatomy, National Taiwan University School of Medicine, Taipei, Taiwan 100, Republic of China a-Internexin is the first neuronal intermediate filament (IF) pro- Behavioral tests demonstrate that these mice have a deficit in tein expressed in postmitotic neurons of the developing ner- motor coordination as early as 3 months of age, consistent with vous system. In the adult, its expression is restricted to mature the morphological neuronal changes. Our data further demon- neurons in the CNS. To study the potential role of a-internexin strate that the neurofilamentous inclusions also lead to progres- in neurodegeneration, we have generated transgenic mice that sive loss of neurons in the aged transgenic mice. The motor overexpress rat a-internexin. The total levels of a-internexin coordination deficit and the loss of neurons are transgene expressed in the hemizygous and homozygous transgenic mice dosage-dependent. These data yield direct evidence that high were ;2 and ;3 times the normal level, respectively. Overex- levels of misaccumulated neuronal IFs lead to neuronal dys- pression of a-internexin resulted in the formation of cerebellar function, progressive neurodegeneration, and ultimate loss of torpedoes as early as 1 month of age. These torpedoes are neurons. -

Role of Gigaxonin in the Regulation of Intermediate Filaments: a Study Using Giant Axonal Neuropathy Patient-Derived Induced Pluripotent Stem Cell-Motor Neurons
Role of Gigaxonin in the Regulation of Intermediate Filaments: a Study Using Giant Axonal Neuropathy Patient-Derived Induced Pluripotent Stem Cell-Motor Neurons Bethany Johnson-Kerner Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy under the Executive Committee of the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2013 © 2012 Bethany Johnson-Kerner All rights reserved Abstract Role of Gigaxonin in the Regulation of Intermediate Filaments: a Study Using Giant Axonal Neuropathy Patient-Derived Induced Pluripotent Stem Cell-Motor Neurons Bethany Johnson-Kerner Patients with giant axonal neuropathy (GAN) exhibit loss of motor and sensory function and typically live for less than 30 years. GAN is caused by autosomal recessive mutations leading to low levels of gigaxonin, a ubiquitously-expressed cytoplasmic protein whose cellular roles are poorly understood. GAN pathology is characterized by aggregates of intermediate filaments (IFs) in multiple tissues. Disorganization of the neuronal intermediate filament (nIF) network is a feature of several neurodegenerative disorders, including amyotrophic lateral sclerosis, Parkinson’s disease and axonal Charcot-Marie-Tooth disease. In GAN such changes are often striking: peripheral nerve biopsies show enlarged axons with accumulations of neurofilaments; so called “giant axons.” Interestingly, IFs also accumulate in other cell types in patients. These include desmin in muscle fibers, GFAP (glial fibrillary acidic protein) in astrocytes, and vimentin in multiple cell types including primary cultures of biopsied fibroblasts. These findings suggest that gigaxonin may be a master regulator of IFs, and understanding its function(s) could shed light on GAN as well as the numerous other diseases in which IFs accumulate. -

Analysis of Protein Expression Regulated by Lumican in PANC‑1 Cells Using Shotgun Proteomics
ONCOLOGY REPORTS 30: 1609-1621, 2013 Analysis of protein expression regulated by lumican in PANC‑1 cells using shotgun proteomics TETSUSHI YAMAMOTO, MITSUHIRO KUDO, WEI-XIA PENG and ZENYA NAITO Departments of Pathology and Integrative Oncological Pathology, Nippon Medical School, Bunkyo-ku, Tokyo 113-8602, Japan Received March 19, 2013; Accepted May 30, 2013 DOI: 10.3892/or.2013.2612 Abstract. Lumican, a member of the class II small leucine- the core protein (1,2). Amino acid sequencing indicates that rich proteoglycan family, regulates the assembly and diameter lumican has 4 potential sites for N-linked keratan sulfate (KS) of collagen fibers in the extracellular matrix of various tissues. or oligosaccharides (3,4). Therefore, lumican includes a core We previously reported that lumican expression in the stromal protein, glycoprotein and proteoglycan forms due to glycosyl- tissues of pancreatic ductal adenocarcinoma (PDAC) correlates ation (5). Lumican is a secreted collagen-binding extracellular with tumor invasion, and tends to correlate with poor prog- matrix protein of the cornea, dermis and tendon stroma, nosis. Lumican stimulates growth and inhibits the invasion of arterial wall, and intestinal submucosa (6-9). Corneal opacity, a PDAC cell line. In the present study, we performed a global as well as skin and tendon fragility due to disorganized and shotgun proteomic analysis using lumican-overexpressing loosely packed collagen fibers in lumican-null mice suggest PANC-1 cells and lumican downregulated PANC-1 cells to that lumican plays an important role in collagen fibrillogen- identify candidate proteins that are regulated by lumican and esis (10,11). related to cell growth and invasion in PDAC cells. -
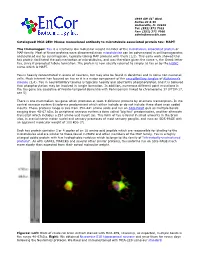
Encor Biotechnology Inc
4949 SW 41st Blvd. Suites 40 & 50 Gainesville, FL 32608 Tel: (352) 372 7022 Fax: (352) 372 7066 [email protected] Catalogue# MCA-2E9: Mouse monoclonal antibody to microtubule associated protein tau- MAPT The Immunogen: Tau is a relatively low molecular weight member of the microtubule associated protein or MAP family. Most of these proteins were discovered since microtubules can be polymerized in cell homogenates and pelleted out by centrifugation, typically taking MAP proteins with them (1,2). This early work showed that tau protein facilitated the polymerization of microtubules, and was therefore given the name τ, the Greek letter tau, since it promoted tubule formation. The protein is now usually referred to simply as tau or by the HGNC name which is MAPT. Tau is heavily concentrated in axons of neurons, but may also be found in dendrites and in some non neuronal cells. Much interest has focused on tau as it is a major component of the neurofibrillary tangles of Alzheimer’s disease (3,4). Tau in neurofibrillary tangles is typically heavily and aberrantly phosphorylated, and it is believed that phosphorylation may be involved in tangle formation. In addition, numerous different point mutations in the tau gene are causative of Fronto-temporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17, see 5). There is one mammalian tau gene which produces at least 9 different proteins by alternate transcription. In the central nervous system 6 isoforms predominant which either include or do not include three short exon coded inserts. These proteins range is size from 352-441 amino acids and run on SDS-PAGE gels as multiple bands ranging from 48-67 kDa.