Borrelia Sp. Phylogenetically Different from Lyme Disease- and Relapsing Fever-Related Borrelia Spp. in Amblyomma Varanense from Python Reticulatus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

First Record & Clinical Management of Tick Infestation by Amblyomma
Int. J. Adv. Res. Biol. Sci. (2020). 7(5): 71-74 International Journal of Advanced Research in Biological Sciences ISSN: 2348-8069 www.ijarbs.com DOI: 10.22192/ijarbs Coden: IJARQG (USA) Volume 7, Issue 5 -2020 Short Communication DOI: http://dx.doi.org/10.22192/ijarbs.2020.07.05.009 First Record & Clinical Management of Tick Infestation by Amblyomma gervaisi, Giardiasis and Tail Injury in a Bengal Monitor (Varanus bengalensis; Daudin, 1802) in Himmatnagar, Gujarat (India) C. M. Bhadesiya*, V. A. Patel, P. J. Gajjar and M. J. Anikar Postgraduate Institute of Veterinary Education & Research (PGIVER), Kamdhenu University, Rajpur (Nava), Himmatnagar - 383010, Gujarat (India) *Corresponding author: [email protected] Abstract A Bengal monitor (Varanus bengalensis; Daudin, 1802) was rescued from a house near Rajpur village of Himmatnagar, Sabarkantha district, Gujarat (India) and brought to the Veterinary Hospital of Kamdhenu University at Rajpur for physical checkup before release. Physical examination revealed minor injury on tail and clinical tick infestation. Ticks were identified as Amblyomma gervaisi while excreta revealed presence of Giardia spp.. The present paper is the first record of Amblyomma gervaisi tick, giardiasis and tail injury in a Bengal monitor in Himmatnagar, Gujarat which will provide baseline information for future research. Keywords: Bengal monitor, Tick, Amblyomma gervaisi, Giardiasis, Gujarat Introduction The Bengal monitor (Varanus bengalensis; Daudin, veterinary case studies in different areas. Some 1802) or a ‘Common Indian Monitor’ is generally relevant publications include [1] Report on Aponomma found in Indian subcontinent including most of the gervaisi as a reptile parasite in Pakistan and India by states. It is included under the ‘Least Concern’ Auffenberg and Auffenberg (1990); [2] Aponomma category by the International Union for Conservation gibsoni tick infestation in monitor lizard at Nagpur by of Nature (IUCN) but the population trend is shown to Harkare et al. -

Aponomma Varanense, of Indian King Cobra Received: 29-09-2016 Accepted: 30-10-2016
Journal of Entomology and Zoology Studies 2016; 4(6): 433-437 E-ISSN: 2320-7078 P-ISSN: 2349-6800 Surface ultra structural Studies of an ectoparasite- JEZS 2016; 4(6): 433-437 © 2016 JEZS Aponomma varanense, of Indian King Cobra Received: 29-09-2016 Accepted: 30-10-2016 Gautam Patra Gautam Patra, Sonjoy Kumar Borthakur, Rajkumari Sunjukta Devi, Department of Veterinary H Lalrinkima and Hmar Lalliankimi Parasitology College of Veterinary Sciences and Animal Husbandry Central Agricultural Abstract University, Selesih, Aizawl, The objective of the present study is to underscore the ultra-structural morphology of Aponomma Mizoram, India varanense-a rare tick of reptile. A. varanense were collected from an Indian Cobra during post mortem and subsequently processed for both light microscopy and Scanning Electron Microscopy (SEM) studies. Sonjoy Kumar Borthakur Light microscopy study revealed few typical morphological features through which the collected Department of Veterinary specimen identification was confirmed. Further, SEM study revealed little morphological ultrastructural Parasitology College of peculiarity with other ixodid ticks as well difference between male and female A. varenense itself. Veterinary Sciences and Animal Occurrence of A. varanense is reported first time from this part of country and this is the first SEM Husbandry Central Agricultural studies of this hard tick from India. University, Selesih, Aizawl, Mizoram, India Keywords: Aponomma, morphology, SEM, Cobra, India Rajkumari Sunjukta Devi Scientist, ICAR, Barapani, 1. Introduction Meghalaya, India Ectoparasites, like ticks and mites are often found on snakes in the wild but the rate of infections and the effects they have on their hosts are poorly studied [1]. Although it is rare that H Lalrinkima ticks occur in large numbers to cause either serious blood loss or other direct injuries but they Department of Veterinary [2, 3] Parasitology College of are responsible for transmission of various vector borne diseases to animals and humans . -

Arthropod Parasites in Domestic Animals
ARTHROPOD PARASITES IN DOMESTIC ANIMALS Abbreviations KINGDOM PHYLUM CLASS ORDER CODE Metazoa Arthropoda Insecta Siphonaptera INS:Sip Mallophaga INS:Mal Anoplura INS:Ano Diptera INS:Dip Arachnida Ixodida ARA:Ixo Mesostigmata ARA:Mes Prostigmata ARA:Pro Astigmata ARA:Ast Crustacea Pentastomata CRU:Pen References Ashford, R.W. & Crewe, W. 2003. The parasites of Homo sapiens: an annotated checklist of the protozoa, helminths and arthropods for which we are home. Taylor & Francis. Taylor, M.A., Coop, R.L. & Wall, R.L. 2007. Veterinary Parasitology. 3rd edition, Blackwell Pub. HOST-PARASITE CHECKLIST Class: MAMMALIA [mammals] Subclass: EUTHERIA [placental mammals] Order: PRIMATES [prosimians and simians] Suborder: SIMIAE [monkeys, apes, man] Family: HOMINIDAE [man] Homo sapiens Linnaeus, 1758 [man] ARA:Ast Sarcoptes bovis, ectoparasite (‘milker’s itch’)(mange mite) ARA:Ast Sarcoptes equi, ectoparasite (‘cavalryman’s itch’)(mange mite) ARA:Ast Sarcoptes scabiei, skin (mange mite) ARA:Ixo Ixodes cornuatus, ectoparasite (scrub tick) ARA:Ixo Ixodes holocyclus, ectoparasite (scrub tick, paralysis tick) ARA:Ixo Ornithodoros gurneyi, ectoparasite (kangaroo tick) ARA:Pro Cheyletiella blakei, ectoparasite (mite) ARA:Pro Cheyletiella parasitivorax, ectoparasite (rabbit fur mite) ARA:Pro Demodex brevis, sebacceous glands (mange mite) ARA:Pro Demodex folliculorum, hair follicles (mange mite) ARA:Pro Trombicula sarcina, ectoparasite (black soil itch mite) INS:Ano Pediculus capitis, ectoparasite (head louse) INS:Ano Pediculus humanus, ectoparasite (body -

Epidemiology of Crimean-Congo Hemorrhagic Fever in Senegal: Temporal and Spatial Patterns
Arch Virol (1990) [Suppl I]: 323-340 0 by Springer-Verlag 1990 Epidemiology of Crimean-Congo hemorrhagic fever in Senegal: temporal and spatial patterns M. L. Wilson''2, J.-P. G>zale~l'~, B. LeGuenno', J.-P. Cornet3, M. Guillaud4, M.-A. Caívo', J.-P. Digoutte', and J.-L. Camicas' c 'Institut Pasteur, Dakar, Senegal 'Departments of Population Sciences and Tropical Public Health, Harvard School of Public Health, Boston, Massachusetts, U.S.A. 31nstitut Francais de Recherche Scientifique pour le Developpement en Cooperation (ORSTOM), Laboratoire ORSTOM de Zoologie medicale, Dakar, Senegal "Institut d'Elevage et de Medecine Veterinaire des Pays Tropicaux, Maisons Alfort, France Accepted April 15, 1990 Summary. Aspects of the spatial and temporal patterns of transmission of Crimean-Congo hemorrhagic fever (CCHF) virus were studied in Senegal, West Africa. A country-wide serological survey of domestic animals indi- cated that transmission was most intense in the northern dry sahelian zone and least in the southern, more humid guinean zone. Human IgG prevalence, ranging from nearly 20% to < 1% among 8 sites throughout the region, also was greatest in the north. A fatal human case of CCHF from Rosso, Mauritania in 1988 was studied and an accompanying serosurvey of human contacts and domestic animals indicated epidemic transmission during that period. Systematic samples of adult ixodid ticks on domestic animals allowed us to analyze the distribution and relative abundance of potential CCHF virus vectors, demonstrating that Hyalomma spp. predominated in those biotopes where transmission was most intense. A prospective study of CCHF virus infection and tick infestation in sheep exposed a period of epizootic transmission in 1988 that corresponded temporally with increased abund- ance of adult H. -

An Investigation on First Outbreak of Kyasanur Forest Disease In
Journal of Entomology and Zoology Studies 2015; 3(6): 239-240 E-ISSN: 2320-7078 P-ISSN: 2349-6800 An investigation on first outbreak of Kyasanur JEZS 2015; 3(6): 239-240 © 2015 JEZS forest disease in Wayanad district of Kerala Received: 18-09-2015 Accepted: 21-10-2015 Prakasan K Prakasan K Abstract Post Graduate and Research As a new challenge to health scenario of Kerala, an outbreak of Kyasanur Forest Disease was reported Dept. of Zoology Maharajas from Wayanad and Malappuram districts of Kerala, India from January to March 2015. A study on tick College Ernakulam Kerala- vectors of Wayanad district confirmed the presence of larval and nymphal Haemaphysalis spinigera, the 682011, India. chief vector of KFD in the forest pastures of Pulppally area. But in other sites viz Kalpetta and Mananthavady its prevalence was found to be low. Presence of tick species like Haemaphysalis bispinosa, Haemaphysalis spinigera, Amblyomma integrum. and Boophilus(Rhipicephalus) annulatus. was also noticed from the host animals surveyed. Keywords: Kyasanur Forest Disease, Ixodid ticks, Ectoparasite, Haemaphysalis Kyasanur Forest Disease (KFD) 1. Introduction Kyasanur Forest Disease (KFD), also referred to as monkey fever is an infectious bleeding disease in monkey and human caused by a highly pathogenic virus called KFD virus. It is an acute prostrating febrile illness, transmitted by infective ticks, especially, Haemaphysalis spinigera. Later KFD viruses also reported from sixteen other species of ticks. Rodents, Shrews, Monkeys and birds upon tick bite become reservoir for this virus. This disease was first reported from the Shimoga district of Karnataka, India in March 1955. Common targets of the virus among monkeys are langur (Semnopithecus entellus) and bonnet monkey (Macaca radiata). -

Amblyomma Hebraeum Is a Hard Tick That Infests Livestock and Wildlife
Amblyomma Importance Amblyomma hebraeum is a hard tick that infests livestock and wildlife. It also hebraeum bites humans. The long mouthparts of Amblyomma ticks make them difficult to remove manually; these ticks also leave large wounds that may become infected by Bont Tick, bacteria or infested by screwworms. A. hebraeum can transmit Ehrlichia ruminantium Southern Africa Bont Tick (formerly Cowdria ruminantium), the agent of heartwater. This tick also carries Rickettsia africae, the agent of African tick-bite fever, an emerging zoonosis in rural sub-Saharan Africa and the Caribbean Last Updated: December 2006 Species Affected Immature A. hebraeum ticks feed on small mammals, ground-feeding birds and reptiles. Adult ticks can be found on livestock and wildlife including antelope. Geographic Distribution A. hebraeum is found in the tropics and subtropics. It prefers moderately humid, warm savannas. This tick is endemic in African countries including South Africa, Zimbabwe, Botswana, Namibia, Malawi, Mozambique and Angola. Life Cycle Amblyomma hebraeum is a three-host tick. Immature ticks feed on small mammals, ground-feeding birds, reptiles and all domestic ruminant species. Adult ticks can be found on livestock and wildlife including antelope, and are usually located on the relatively hairless parts of the body. Most are found on the ventral body surface, the perineum, and the axillae, as well as under the tail. Identification A. hebraeum is a member of the family Ixodidae (hard ticks). Hard ticks have a dorsal shield (scutum) and their mouthparts (capitulum) protrude forward when they are seen from above. Amblyomma ticks are large variegated ticks with long, strong mouthparts. -

Investigations on the Abundance of Ectoparasites and Vector-Borne Pathogens in Southwest Madagascar
Investigations on the abundance of ectoparasites and vector-borne pathogens in southwest Madagascar Dissertation with the aim of achieving a doctoral degree at the Faculty of Mathematics, Informatics and Natural Sciences Department of Biology University of Hamburg submitted by Julian Ehlers Hamburg, 2020 Reviewers: Prof. Dr. Jörg Ganzhorn, Universität Hamburg PD Dr. Andreas Krüger, Centers for Disease Control and Prevention Date of oral defense: 19.06.2020 TABLE OF CONTENTS Table of contents Summary 1 Zusammenfassung 3 Chapter 1: General introduction 5 Chapter 2: Ectoparasites of endemic and domestic animals in 33 southwest Madagascar Chapter 3: Molecular detection of Rickettsia spp., Borrelia spp., 63 Bartonella spp. and Yersinia pestis in ectoparasites of endemic and domestic animals in southwest Madagascar Chapter 4: General discussion 97 SUMMARY Summary Human encroachment on natural habitats is steadily increasing due to the rapid growth of the worldwide population. The consequent expansion of agricultural land and livestock husbandry, accompanied by spreading of commensal animals, create new interspecific contact zones that are major regions of risk of the emergence of diseases and their transmission between livestock, humans and wildlife. Among the emerging diseases of the recent years those that originate from wildlife reservoirs are of outstanding importance. Many vector-borne diseases are still underrecognized causes of fever throughout the world and tend to be treated as undifferentiated illnesses. The lack of human and animal health facilities, common in rural areas, bears the risk that vector-borne infections remain unseen, especially if they are not among the most common. Ectoparasites represent an important route for disease transmission besides direct contact to infected individuals. -
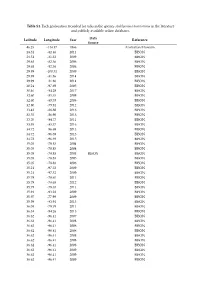
Table S1. Each Geolocation Recorded for Ticks in the Species Amblyomma Americanum in the Literature and Publicly Available Online Databases
Table S1. Each geolocation recorded for ticks in the species Amblyomma americanum in the literature and publicly available online databases. Data Latitude Longitude Year Reference Source 46.25 -114.17 1966 Australian Museum 28.31 -82.46 2011 BISON 28.51 -81.32 2009 BISON 29.68 -82.36 2006 BISON 29.68 -82.36 2006 BISON 29.99 -100.31 2009 BISON 29.99 -81.86 2014 BISON 29.99 -81.86 2014 BISON 30.24 -97.69 2005 BISON 30.46 -84.28 2017 BISON 32.60 -85.35 2008 BISON 32.60 -85.35 2008 BISON 32.80 -79.94 2012 BISON 33.42 -88.88 2016 BISON 33.55 -86.90 2013 BISON 33.70 -84.77 2011 BISON 33.95 -83.37 2016 BISON 34.72 -96.69 2013 BISON 34.72 -96.69 2013 BISON 34.72 -96.69 2015 BISON 35.05 -78.83 2004 BISON 35.05 -78.83 2004 BISON 35.05 -78.83 2004 BISON BISON 35.05 -78.83 2005 BISON 35.05 -78.83 2006 BISON 35.21 -97.32 2009 BISON 35.21 -97.32 2009 BISON 35.79 -78.65 2011 BISON 35.79 -78.65 2012 BISON 35.79 -78.65 2011 BISON 35.91 -93.22 2009 BISON 35.97 -77.99 2009 BISON 35.99 -83.94 2015 BISON 36.08 -79.79 2011 BISON 36.34 -94.26 2015 BISON 36.62 -96.41 2007 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2008 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2009 BISON 36.62 -96.41 2011 BISON 36.62 -96.41 2012 BISON 36.62 -96.41 2012 BISON 37.36 -77.05 2014 BISON 37.62 -84.87 2016 BISON 37.83 -78.28 2011 BISON 37.97 -85.70 2016 BISON 37.97 -85.70 2016 BISON 38.22 -75.31 2014 BISON 38.43 -88.43 -

Amblyomma Variegatum S L I D E 1 Amblyomma Variegatum Is a Hard
Amblyomma variegatum S Amblyomma variegatum is a hard tick that feeds on a number of l domesticated animals including cattle, sheep, goats, horses and dogs, as i well as humans. The long mouthparts of A. variegatum leave large wounds, and make this tick difficult to remove manually. Its bite is d Amblyomma variegatum e severe and painful, and can result in significant damage to the skin. Secondary infections can cause septic wounds or abscesses, and Tropical Bont Tick 1 Tropical African Bont Tick inflammation on the teats of cows may affect milk production. In some regions, Amblyomma bite wounds may become infested by screwworms. In addition, A. variegatum is a host for a number of microbial pathogens including Ehrlichia ruminantium (formerly Cowdria ruminantium), the agent of heartwater, and Rickettsia africae, the agent of African tick-bite fever, which is an emerging zoonosis in rural sub-Saharan Africa and the Caribbean. S In today’s presentation we will cover information regarding the tick l Overview Amblyomma variegatum and the diseases it can transmit. We will also i • Organism talk about how to identify the tick, and the impact this tick has had in the d • Identification past and could have in the future. Additionally, we will talk about how it • Importance is transmitted and the species it affects. Finally, we will address e • Geographic Distribution • Life Cycle prevention and control measures, as well as actions to take if • Associated Diseases Amblyomma variegatum is suspected. 2 • Prevention and Control • Recommended Actions Center for Food Security and Public Health, Iowa State University, 2011 S Amblyomma variegatum is a hard tick in the family Ixodidae. -

Candidatus Rickettsia Colombianensi in Ticks from Reptiles in Córdoba, Colombia
Veterinary World, EISSN: 2231-0916 RESEARCH ARTICLE Available at www.veterinaryworld.org/Vol.13/September-2020/5.pdf Open Access Candidatus Rickettsia colombianensi in ticks from reptiles in Córdoba, Colombia Jorge Miranda1 , Lina Violet-Lozano1 , Samia Barrera1 , Salim Mattar1 , Santiago Monsalve-Buriticá2 , Juan Rodas3 and Verónica Contreras1 1. University of Córdoba, Institute of Tropical Biology Research, Córdoba, Colombia; 2. Corporación Universitaria Lasallista, Colombia; 3. University of Antioquia, Colombia, Colombia. Corresponding author: Jorge Miranda, e-mail: [email protected] Co-authors: LV: [email protected], SB: [email protected], SM: [email protected], SaM: [email protected], JR: [email protected], VC: [email protected] Received: 11-02-2020, Accepted: 09-07-2020, Published online: 03-09-2020 doi: www.doi.org/10.14202/vetworld.2020.1764-1770 How to cite this article: Miranda J, Violet-Lozano L, Barrera S, Mattar S, Monsalve-Buriticá S, Rodas J, Contreras V (2020) Candidatus Rickettsia colombianensi in ticks from reptiles in Córdoba, Colombia, Veterinary World, 13(9): 1764-1770. Abstract Background and Aim: Wildlife animals are reservoirs of a large number of microorganisms pathogenic to humans, and ticks could be responsible for the transmission of these pathogens. Rickettsia spp. are the most prevalent pathogens found in ticks. This study was conducted to detect Rickettsia spp. in ticks collected from free-living and illegally trafficked reptiles from the Department of Córdoba, Colombia. Materials and Methods: During the period from October 2011 to July 2014, ticks belonging to the family Ixodidae were collected, preserved in 96% ethanol, identified using taxonomic keys, and pooled (between 1 and 14 ticks) according to sex, stage, host, and collected place for subsequent DNA extraction. -

Species Composition of Hard Ticks (Acari: Ixodidae) on Domestic Animals and Their Public Health Importance in Tamil Nadu, South India
Acarological Studies Vol 3 (1): 16-21 doi: 10.47121/acarolstud.766636 RESEARCH ARTICLE Species composition of hard ticks (Acari: Ixodidae) on domestic animals and their public health importance in Tamil Nadu, South India Krishnamoorthi RANGANATHAN1 , Govindarajan RENU2 , Elango AYYANAR1 , Rajamannar VEERAMANO- HARAN2 , Philip Samuel PAULRAJ2,3 1 ICMR-Vector Control Research Centre, Puducherry, India 2 ICMR-Vector Control Research Centre Field Station, Madurai, Tamil Nadu, India 3 Corresponding author: [email protected] Received: 8 July 2020 Accepted: 4 November 2020 Available online: 27 January 2021 ABSTRACT: This study was carried out in Madurai district, Tamil Nadu State, South India. Ticks were collected from cows, dogs, goats, cats and fowls. The overall percentage of tick infestation in these domestic animals was 21.90%. The following ticks were identified: Amblyomma integrum, Haemaphysalis bispinosa, Haemaphysalis paraturturis, Haemaphy- salis turturis, Haemaphysalis intermedia, Haemaphysalis spinigera, Hyalomma anatolicum, Hyalomma brevipunctata, Hy- alomma kumari, Rhipicephalus turanicus, Rhipicephalus haemaphysaloides and Rhipicephalus sanguineus. The predomi- nant species recorded from these areas is R. sanguineus (27.03%) followed by both R (B.) microplus (24.12%) and R. (B.) decoloratus (18.82%). The maximum tick infestation rate was recorded in animals from rural areas (25.67%), followed by semi-urban (21.66%) and urban (16.05%) areas. This study proved the predominance of hard ticks as parasites on domestic animals and will help the public health personnel to understand the ground-level situation and to take up nec- essary control measures to prevent tick-borne diseases. Keywords: Ticks, domestic animals, Ixodidae, prevalence. Zoobank: http://zoobank.org/D8825743-B884-42E4-B369-1F16183354C9 INTRODUCTION longitude is 78.0195° E. -

The Prevalence of the Q-Fever Agent Coxiella Burnetii in Ticks Collected from an Animal Shelter in Southeast Georgia
Georgia Southern University Digital Commons@Georgia Southern Electronic Theses and Dissertations Graduate Studies, Jack N. Averitt College of Summer 2004 The Prevalence of the Q-fever Agent Coxiella burnetii in Ticks Collected from an Animal Shelter in Southeast Georgia John H. Smoyer III Follow this and additional works at: https://digitalcommons.georgiasouthern.edu/etd Part of the Immunology of Infectious Disease Commons, Other Animal Sciences Commons, and the Parasitology Commons Recommended Citation Smoyer, John H. III, "The Prevalence of the Q-fever Agent Coxiella burnetii in Ticks Collected from an Animal Shelter in Southeast Georgia" (2004). Electronic Theses and Dissertations. 1002. https://digitalcommons.georgiasouthern.edu/etd/1002 This thesis (open access) is brought to you for free and open access by the Graduate Studies, Jack N. Averitt College of at Digital Commons@Georgia Southern. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Digital Commons@Georgia Southern. For more information, please contact [email protected]. THE PREVALENCE OF THE Q-FEVER AGENT COXIELLA BURNETII IN TICKS COLLECTED FROM AN ANIMAL SHELTER IN SOUTHEAST GEORGIA by JOHN H. SMOYER, III (Under the Direction of Quentin Q. Fang) ABSTRACT Q-fever is a zoonosis caused by a worldwide-distributed bacterium Coxiella burnetii . Ticks are vectors of the Q-fever agent but play a secondary role in transmission because the agent is also transmitted via aerosols. Most Q-fever studies have focused on farm animals but not ticks collected from dogs in animal shelters. In order to detect the Q-fever agent in these ticks, a nested PCR technique targeting the 16S rDNA of Coxiella burnetii was used.