The ARID Domain Protein Dril1 Is Necessary for Tgfh Signaling in Xenopus Embryos$
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization and Stress Response of the Jmjc Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium Hirsutum
plants Article Characterization and Stress Response of the JmjC Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium hirsutum 1, 2, 2, 3 3 3 Jie Zhang y, Junping Feng y, Wei Liu *, Zhongying Ren , Junjie Zhao , Xiaoyu Pei , Yangai Liu 3, Daigang Yang 3 and Xiongfeng Ma 1,3,* 1 Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China; [email protected] 2 Collaborative Innovation Center of Henan Grain Crops, Agronomy College, Henan Agricultural University, Zhengzhou 450002, China; [email protected] 3 State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang 455000, China; [email protected] (Z.R.); [email protected] (J.Z.); [email protected] (X.P.); [email protected] (Y.L.); [email protected] (D.Y.) * Correspondence: [email protected] (W.L.); [email protected] (X.M.) These authors contributed equally to this work. y Received: 12 October 2020; Accepted: 18 November 2020; Published: 20 November 2020 Abstract: Histone modification is an important epigenetic modification that controls gene transcriptional regulation in eukaryotes. Histone methylation is accomplished by histone methyltransferase and can occur on two amino acid residues, arginine and lysine. JumonjiC (JmjC) domain-containing histone demethylase regulates gene transcription and chromatin structure by changing the methylation state of the lysine residue site and plays an important role in plant growth and development. In this study, we carried out genome-wide identification and comprehensive analysis of JmjC genes in the allotetraploid cotton species Gossypium hirsutum. In total, 50 JmjC genes were identified and in G. -

Associating Regions Are Bound by Bright: a B Cell-Specific Trans-Activator That Describes a New DNA-Binding Protein Family
Downloaded from genesdev.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press The immunoglobulin.. heavy-chain matrix-. associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family Richard F. Herrscher, 1,4 Mark H. Kaplan, 1'3 David L. Lelsz, 1 Chhaya Das, 1'4 Richard Scheuermann, 2 and Philip W. Tucker 1'4'5 Departments of ~Microbiology, 2Pathology, and the Immunology Graduate Program, University of Texas Southwestern Medical Center, Dallas, Texas 75235-9048 USA B lymphocyte-restricted transcription of immunoglobulin heavy-chain (IgH) genes is specified by elements within the variable region (Va) promoter and the intronic enhancer (EIX). The gene encoding a protein that binds a VH promoter proximal site necessary for induced ix-heavy-chain transcription has been cloned. This B-cell specific protein, termed Bright (B cell regulator of IgH transcription), is found in both soluble and matrix insoluble nuclear fractions. Bright binds the minor groove of a restricted ATC sequence that is sufficient for nuclear matrix association. This sequence motif is present in previously described matrix-associating regions (MARs) proximal to the promoter and flanking EIX. Bright can activate Ela-driven transcription by binding these sites, but only when they occur in their natural context and in cell lines permissive for E~ activity. To bind DNA, Bright requires a novel tetramerization domain and a previously undescribed domain that shares identity with several proteins, including SWI1, a component of the SWI/SNF complex. [Key Words: Nuclear matrix; matrix-associating regions; MAR-binding protein; immunoglobulin transcription; IgH enhancer; B lymphocytel Received July 14, 1995; revised version accepted October 23, 1995. -

Jarid2 and PRC2, Partners in Regulating Gene Expression
Downloaded from genesdev.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Jarid2 and PRC2, partners in regulating gene expression Gang Li,1,2,6 Raphael Margueron,2,6 Manching Ku,1,3,4 Pierre Chambon,5 Bradley E. Bernstein,1,3,4 and Danny Reinberg1,2,7 1Howard Hughes Medical Institute, New York University Medical School, New York, New York 10016, USA; 2Department of Biochemistry, New York University Medical School, New York, New York 10016, USA; 3Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, Massachusetts 02142, USA; 4Department of Pathology, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts 02114, USA; 5Department of Functional Genomics, Institut de Ge´ne´tique et de Biologie Mole´culaire et Cellulaire (IGBMC), Institut Clinique de la Souris (ICS), CNRS/INSERM/Universite´ de Strasbourg, BP10142, 67404 Illkirch, France The Polycomb group proteins foster gene repression profiles required for proper development and unimpaired adulthood, and comprise the components of the Polycomb-Repressive Complex 2 (PRC2) including the histone H3 Lys 27 (H3K27) methyltransferase Ezh2. How mammalian PRC2 accesses chromatin is unclear. We found that Jarid2 associates with PRC2 and stimulates its enzymatic activity in vitro. Jarid2 contains a Jumonji C domain, but is devoid of detectable histone demethylase activity. Instead, its artificial recruitment to a promoter in vivo resulted in corecruitment of PRC2 with resultant increased levels of di- and trimethylation of H3K27 (H3K27me2/3). Jarid2 colocalizes with Ezh2 and MTF2, a homolog of Drosophila Pcl, at endogenous genes in embryonic stem (ES) cells. Jarid2 can bind DNA and its recruitment in ES cells is interdependent with that of PRC2, as Jarid2 knockdown reduced PRC2 at its target promoters, and ES cells devoid of the PRC2 component EED are deficient in Jarid2 promoter access. -

The Emerging Role of KDM5A in Human Cancer
Yang et al. J Hematol Oncol (2021) 14:30 https://doi.org/10.1186/s13045-021-01041-1 REVIEW Open Access The emerging role of KDM5A in human cancer Guan‑Jun Yang1,2,3,4 , Ming‑Hui Zhu1,2,3, Xin‑Jiang Lu1,2,3, Yan‑Jun Liu5, Jian‑Fei Lu1,2,3, Chung‑Hang Leung4*, Dik‑Lung Ma6* and Jiong Chen1,2,3* Abstract Histone methylation is a key posttranslational modifcation of chromatin, and its dysregulation afects a wide array of nuclear activities including the maintenance of genome integrity, transcriptional regulation, and epigenetic inherit‑ ance. Variations in the pattern of histone methylation infuence both physiological and pathological events. Lysine‑ specifc demethylase 5A (KDM5A, also known as JARID1A or RBP2) is a KDM5 Jumonji histone demethylase subfamily member that erases di‑ and tri‑methyl groups from lysine 4 of histone H3. Emerging studies indicate that KDM5A is responsible for driving multiple human diseases, particularly cancers. In this review, we summarize the roles of KDM5A in human cancers, survey the feld of KDM5A inhibitors including their anticancer activity and modes of action, and the current challenges and potential opportunities of this feld. Keywords: KDM5A, Cancer, Jumonji C domain, Histone methylation, Drug resistance, Targeted therapy Background NUP98 and KDM5A, mediates hematopoietic cell prolif- Lysine-specifc demethylase 5A (KDM5A), also named eration and alters myelo-erythropoietic diferentiation via Jumonji/ARID domain-containing protein 1A (JARID1A) demethylating H3K4me2/3 [14–17]. In terms of mecha- or retinoblastoma-binding protein 2 (RBP2), origi- nism, KDM5A and its fusion gene Fe(II)-dependently nally reported as a retinoblastoma protein (RB) pocket catalyzes oxidative decarboxylation of 2OG with con- domain-binding protein in 2001 [1], is a Fe(II)- and sumption of O2 to generate a reactive iron(IV)-oxo inter- α-ketoglutaric acid (2OG)-dependent JmjC-containing mediate, carbon dioxide, and succinate. -

The Histone Demethylase KDM5 Is Essential for Larval Growth in Drosophila
bioRxiv preprint doi: https://doi.org/10.1101/297804; this version posted April 9, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The histone demethylase KDM5 is essential for larval growth in Drosophila Coralie Drelon, Helen M. Belalcazar, and Julie Secombe1 Department of Genetics Albert Einstein College of Medicine 1300 Morris Park Avenue Bronx, NY, 10461 Phone (718) 430 2698 1 corresponding author: [email protected] Running title: KDM5 regulates larval growth rate Key words: KDM5, Lid, H3K4me3, chromatin, growth, imaginal disc 1 bioRxiv preprint doi: https://doi.org/10.1101/297804; this version posted April 9, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Regulated gene expression is necessary for developmental and homeostatic processes. The KDM5 family of proteins are histone H3 lysine 4 demethylases that can regulate transcription through both demethylase-dependent and independent mechanisms. While loss and overexpression of KDM5 proteins are linked to intellectual disability and cancer, respectively, their normal developmental functions remain less characterized. Drosophila melanogaster provides an ideal system to investigate KDM5 function, as it encodes a single ortholog in contrast to the four paralogs found in mammalian cells. To examine the consequences of complete loss of KDM5, we generated a null allele of Drosophila kdm5, also known as little imaginal discs (lid), and show that it is essential for development. -

ARID2 Chromatin Remodeler in Hepatocellular Carcinoma
cells Review ARID2 Chromatin Remodeler in Hepatocellular Carcinoma Robin Loesch 1,2, Linda Chenane 1,2 and Sabine Colnot 1,2,* 1 INSERM, Centre de Recherche des Cordeliers (CRC), Sorbonne Université, Université de Paris, F-75006 Paris, France; [email protected] (R.L.); [email protected] (L.C.) 2 Equipe labellisée “Ligue Nationale Contre le Cancer”, F-75013 Paris, France * Correspondence: [email protected] Received: 30 July 2020; Accepted: 16 September 2020; Published: 23 September 2020 Abstract: Chromatin remodelers are found highly mutated in cancer including hepatocellular carcinoma. These mutations frequently occur in ARID (AT-rich Interactive Domain) genes, encoding subunits of the ATP-dependent SWI/SNF remodelers. The increasingly prevalent complexity that surrounds the functions and specificities of the highly modular BAF (BG1/BRM-associated factors) and PBAF (polybromo-associated BAF) complexes, including ARID1A/B or ARID2, is baffling. The involvement of the SWI/SNF complexes in diverse tissues and processes, and especially in the regulation of gene expression, multiplies the specific outcomes of specific gene alterations. A better understanding of the molecular consequences of specific mutations impairing chromatin remodelers is needed. In this review, we summarize what we know about the tumor-modulating properties of ARID2 in hepatocellular carcinoma. Keywords: ARID2; hepatocellular carcinoma; cancer; chromatin; SWI/SNF 1. Introduction to the SWI/SNF Chromatin Remodeling Complexes Gene expression regulation is far from being a random phenomenon. It requires covalent modifications targeting both DNA via methylation, and histones via post-translational modifications (PTM), which shape the nucleosomes and control the DNA compaction into chromatin and the recruitment of specific transcription factors. -

The Many Roles of BAF (Mswi/SNF) and PBAF Complexes in Cancer
Downloaded from http://perspectivesinmedicine.cshlp.org/ at Cold Spring Harbor Laboratory Library on July 14, 2016 - Published by Cold Spring Harbor Laboratory Press The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer Courtney Hodges, Jacob G. Kirkland, and Gerald R. Crabtree Departments of Pathology, Developmental Biology, and Genetics, Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, California 94305 Correspondence: [email protected] During the last decade, a host of epigenetic mechanisms were found to contribute to cancer and other human diseases. Several genomic studies have revealed that 20% of malignan- cies have alterations of the subunits of polymorphic BRG-/BRM-associated factor (BAF) and Polybromo-associated BAF (PBAF) complexes, making them among the most frequently mutated complexes in cancer. Recurrent mutations arise in genes encoding several BAF/ PBAF subunits, including ARID1A, ARID2, PBRM1, SMARCA4, and SMARCB1. These sub- units share some degree of conservation with subunits from related adenosine triphosphate (ATP)-dependent chromatin remodeling complexes in model organisms, in which a large body of work provides insight into their roles in cancer. Here, we review the roles of BAF- and PBAF-like complexes in these organisms, and relate these findings to recent discoveries in cancer epigenomics. We review several roles of BAF and PBAF complexes in cancer, includ- ing transcriptional regulation, DNA repair, and regulation of chromatin architecture and topology.More recent results highlight the need for new techniques to study these complexes. EPIGENOMICS IN CANCER the full spectrum of achievable cell-type diver- sity for the organism. Because epigenetic regu- roadly defined, epigenetic factors contrib- lation contributes to cell-type functional spe- Bute to the expression state of the genome cialization, it is essential for multicellular life. -

ARID1A Gene AT-Rich Interaction Domain 1A
ARID1A gene AT-rich interaction domain 1A Normal Function The ARID1A gene provides instructions for making a protein that forms one piece ( subunit) of several different SWI/SNF protein complexes. SWI/SNF complexes regulate gene activity (expression) by a process known as chromatin remodeling. Chromatin is the network of DNA and protein that packages DNA into chromosomes. The structure of chromatin can be changed (remodeled) to alter how tightly DNA is packaged. Chromatin remodeling is one way gene expression is regulated during development; when DNA is tightly packed, gene expression is lower than when DNA is loosely packed. Through their ability to regulate gene activity, SWI/SNF complexes are involved in many processes, including repairing damaged DNA; copying (replicating) DNA; and controlling the growth, division, and maturation (differentiation) of cells. The ARID1A protein and other SWI/SNF subunits are thought to act as tumor suppressors, which keep cells from growing and dividing too rapidly or in an uncontrolled way. The ARID1A subunit is able to attach (bind) to DNA and is thought to help target SWI/ SNF complexes to the chromatin location that needs to be remodeled. Health Conditions Related to Genetic Changes Coffin-Siris syndrome More than 30 variants (also known as mutations) in the ARID1A gene can cause Coffin- Siris syndrome. This condition is characterized by delayed development, abnormalities of the fifth (pinky) fingers or toes, and characteristic facial features that are described as coarse. The ARID1A gene variants involved in Coffin-Siris syndrome lead to an abnormally short, nonfunctional protein. As a result, affected individuals have half the normal amount of functioning ARID1A protein. -

Transcriptional and Epigenetic Substrates of Methamphetamine Addiction and Withdrawal: Evidence from a Long-Access Self-Administration Model in the Rat
Mol Neurobiol (2015) 51:696–717 DOI 10.1007/s12035-014-8776-8 Transcriptional and Epigenetic Substrates of Methamphetamine Addiction and Withdrawal: Evidence from a Long-Access Self-Administration Model in the Rat Jean Lud Cadet & Christie Brannock & Subramaniam Jayanthi & Irina N. Krasnova Received: 6 March 2014 /Accepted: 1 June 2014 /Published online: 18 June 2014 # The Author(s) 2014. This article is published with open access at Springerlink.com Abstract Methamphetamine use disorder is a chronic neuro- These transcriptional changes might serve as triggers psychiatric disorder characterized by recurrent binge episodes, for the neuropsychiatric presentations of humans who intervals of abstinence, and relapses to drug use. Humans abuse this drug. Better understanding of the way that addicted to methamphetamine experience various degrees of gene products interact to cause methamphetamine addic- cognitive deficits and other neurological abnormalities that tion will help to develop better pharmacological treat- complicate their activities of daily living and their participa- ment of methamphetamine addicts. tion in treatment programs. Importantly, models of metham- phetamine addiction in rodents have shown that animals will Keywords Gene expression . Gene networks . Transcription readily learn to give themselves methamphetamine. Rats also factors . Epigenetics . HDAC . Repressor complexes . accelerate their intake over time. Microarray studies have also Cognition . Striatum shown that methamphetamine taking is associated with major transcriptional -

The Drosophila Retained/Dead Ringer Gene and ARID Gene Family Function During Development
Int. J. Dev. Biol. 46: 423-430 (2002) The Drosophila retained/dead ringer gene and ARID gene family function during development TETYANA SHANDALA1, R. DANIEL KORTSCHAK1 and ROBERT SAINT1,2,* 1Centre for the Molecular Genetics of Development and Dept. of Molecular Biosciences, Adelaide University, Adelaide, SA, Australia and 2Research School of Biological Sciences, Australian National University, Canberra, ACT, Australia ABSTRACT The recently discovered ARID family of proteins interact with DNA through a phylo- genetically conserved sequence termed the A/T Interaction Domain (ARID). The retained/dead ringer (retn/dri) gene of Drosophila melanogaster is a founding member of the ARID gene family, and of the eARID subfamily. This subfamily exhibits an extended region of sequence similarity beyond the core ARID motif and a separate conserved domain termed the REKLES domain. retn/ dri is involved in a range of developmental processes, including axis patterning and muscle development. The retn/dri ARID motif has been shown by in vitro studies to exhibit sequence- specific DNA binding activity. Here we demonstrate that the ARID domain is essential for the in vivo function of retn/dri during embryonic development by showing that a mutant form of RETN/DRI, deleted for part of the ARID domain and unable to bind DNA in vitro, cannot rescue the retn/dri mutant phenotype. In the presence of wild-type RETN/DRI this construct acts as a dominant negative, providing additional support for the proposal that RETN/DRI acts in a multiprotein complex. In contrast, we are yet to find an in vivo role for the REKLES domain, despite its clear evolutionary conservation. -

ARID1B Gene AT-Rich Interaction Domain 1B
ARID1B gene AT-rich interaction domain 1B Normal Function The ARID1B gene provides instructions for making a protein that forms one piece ( subunit) of several different SWI/SNF protein complexes. SWI/SNF complexes regulate gene activity (expression) by a process known as chromatin remodeling. Chromatin is the network of DNA and proteins that packages DNA into chromosomes. The structure of chromatin can be changed (remodeled) to alter how tightly DNA is packaged. Chromatin remodeling is one way gene expression is regulated during development; when DNA is tightly packed, gene expression is lower than when DNA is loosely packed. Through their ability to regulate gene activity, SWI/SNF complexes are involved in many processes, including repairing damaged DNA; copying (replicating) DNA; and controlling the growth, division, and maturation (differentiation) of cells. The ARID1B protein and other SWI/SNF subunits are thought to act as tumor suppressors, which keep cells from growing and dividing too rapidly or in an uncontrolled way. The ARID1B subunit is able to attach (bind) to DNA and is thought to help target SWI/ SNF complexes to the chromatin location that needs to be remodeled. Health Conditions Related to Genetic Changes Coffin-Siris syndrome More than 150 variants (also known as mutations) in the ARID1B gene have been found to cause Coffin-Siris syndrome. This condition is characterized by delayed development, abnormalities of the fifth (pinky) fingers or toes, and characteristic facial features that are described as coarse. Most ARID1B gene variants involved in Coffin-Siris syndrome lead to an abnormally short, nonfunctional protein. As a result, affected individuals have half the normal amount of functional ARID1B protein. -
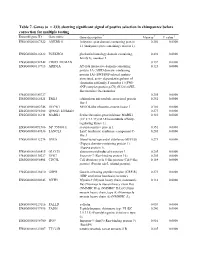
Table 7. Genes
Table 7. Genes (n = 233) showing significant signal of postive selection in chimpanzee before correction for multiple testing Ensembl gene ID Gene name Gene description * Mean ω † P value ‡ ENSG00000167522 ANKRD11 Ankyrin repeat domain-containing protein 0.256 0.0000 11 (Ankyrin repeat-containing cofactor 1). ENSG00000126822 PLEKHG3 pleckstrin homology domain containing, 0.492 0.0000 family G, member 3 ENSG00000152242 CR025_HUMAN 0.137 0.0000 ENSG00000117713 ARID1A AT-rich interactive domain-containing 0.123 0.0000 protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix- associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI- SNF complex protein p270) (B120) (SWI- like protein) (Osa homolog ENSG00000185727 0.205 0.0000 ENSG00000165521 EML5 echinoderm microtubule associated protein 0.252 0.0000 like 5 ENSG00000002746 HECW1 NEDD4-like ubiquitin-protein ligase 1 0.186 0.0000 ENSG00000198308 Q9NSI3_HUMAN 0.273 0.0000 ENSG00000116141 MARK1 Serine/threonine-protein kinase MARK1 0.410 0.0000 (EC 2.7.1.37) (MAP/microtubule affinity- regulating kinase 1). ENSG00000091986 NP_955806.1 steroid-sensitive protein 1 0.351 0.0000 ENSG00000147036 LANCL3 LanC lantibiotic synthetase component C- 0.205 0.0000 like 3 ENSG00000112276 BVES Blood vessel epicardial substance (hBVES) 0.273 0.0000 (Popeye domain-containing protein 1) (Popeye protein 1). ENSG00000106415 GLCCI1 glucocorticoid induced transcript 1 0.205 0.0000 ENSG00000130227 XPO7 Exportin-7 (Ran-binding protein 16). 0.205 0.0000 ENSG00000096401 CDC5L Cell division cycle 5-like protein (Cdc5-like 0.164 0.0000 protein) (Pombe cdc5- related protein). ENSG00000126010 GRPR Gastrin-releasing peptide receptor (GRP-R) 0.273 0.0000 (GRP-preferring bombesin receptor).