Author Index
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Building Affordance Relations for Robotic Agents - a Review
Proceedings of the Thirtieth International Joint Conference on Artificial Intelligence (IJCAI-21) Survey Track Building Affordance Relations for Robotic Agents - A Review Paola Ardon´ , Eric` Pairet , Katrin S. Lohan , Subramanian Ramamoorthy and Ronald P. A. Petrick Edinburgh Centre for Robotics Edinburgh, Scotland, United Kingdom [email protected] Abstract practical implementations of affordances, with applications to tasks such as action prediction, navigation and manipulation. Affordances describe the possibilities for an agent The idea of affordances has been studied from different to perform actions with an object. While the sig- perspectives. Early surveys [Chemero and Turvey, 2007; nificance of the affordance concept has been previ- S¸ahin et al., 2007; Horton et al., 2012] summarise formalisms ously studied from varied perspectives, such as psy- that attempt to bridge the controversial concept of affordances chology and cognitive science, these approaches in psychology with mathematical representations. Other sur- are not always sufficient to enable direct transfer, veys discuss the connection of robotic affordances with other in the sense of implementations, to artificial intel- disciplines [Jamone et al., 2016], and propose classification ligence (AI)-based systems and robotics. However, schemes to review and categorise the related literature [Min many efforts have been made to pragmatically em- et al., 2016; Zech et al., 2017]. In contrast, we focus more ploy the concept of affordances, as it represents on the implications of different design decisions regarding great potential for AI agents to effectively bridge task abstraction and learning techniques that could scale up perception to action. In this survey, we review and in physical domains, to address the need for generalisation in find common ground amongst different strategies the AI sense. -

Marathon 2,500 Years Edited by Christopher Carey & Michael Edwards
MARATHON 2,500 YEARS EDITED BY CHRISTOPHER CAREY & MICHAEL EDWARDS INSTITUTE OF CLASSICAL STUDIES SCHOOL OF ADVANCED STUDY UNIVERSITY OF LONDON MARATHON – 2,500 YEARS BULLETIN OF THE INSTITUTE OF CLASSICAL STUDIES SUPPLEMENT 124 DIRECTOR & GENERAL EDITOR: JOHN NORTH DIRECTOR OF PUBLICATIONS: RICHARD SIMPSON MARATHON – 2,500 YEARS PROCEEDINGS OF THE MARATHON CONFERENCE 2010 EDITED BY CHRISTOPHER CAREY & MICHAEL EDWARDS INSTITUTE OF CLASSICAL STUDIES SCHOOL OF ADVANCED STUDY UNIVERSITY OF LONDON 2013 The cover image shows Persian warriors at Ishtar Gate, from before the fourth century BC. Pergamon Museum/Vorderasiatisches Museum, Berlin. Photo Mohammed Shamma (2003). Used under CC‐BY terms. All rights reserved. This PDF edition published in 2019 First published in print in 2013 This book is published under a Creative Commons Attribution-NonCommercial- NoDerivatives (CC-BY-NC-ND 4.0) license. More information regarding CC licenses is available at http://creativecommons.org/licenses/ Available to download free at http://www.humanities-digital-library.org ISBN: 978-1-905670-81-9 (2019 PDF edition) DOI: 10.14296/1019.9781905670819 ISBN: 978-1-905670-52-9 (2013 paperback edition) ©2013 Institute of Classical Studies, University of London The right of contributors to be identified as the authors of the work published here has been asserted by them in accordance with the Copyright, Designs and Patents Act 1988. Designed and typeset at the Institute of Classical Studies TABLE OF CONTENTS Introductory note 1 P. J. Rhodes The battle of Marathon and modern scholarship 3 Christopher Pelling Herodotus’ Marathon 23 Peter Krentz Marathon and the development of the exclusive hoplite phalanx 35 Andrej Petrovic The battle of Marathon in pre-Herodotean sources: on Marathon verse-inscriptions (IG I3 503/504; Seg Lvi 430) 45 V. -

Livability Court Records 1/1/1997 to 8/31/2021
Livability Court Records 1/1/1997 to 8/31/2021 Last First Middle Case Charge Disposition Disposition Date Judge 133 Cannon St Llc Rep JohnCompany Q Florence U43958 Minimum Standards For Vacant StructuresGuilty 8/13/18 Molony 148 St Phillips St Assoc.Company U32949 Improper Disposal of Garbage/Trash Guilty- Residential 10/17/11 Molony 18 Felix Llc Rep David BevonCompany U34794 Building Permits; Plat and Plans RequiredGuilty 8/13/18 Mendelsohn 258 Coming Street InvestmentCompany Llc Rep Donald Mitchum U42944 Public Nuisances Prohibited Guilty 12/18/17 Molony 276 King Street Llc C/O CompanyDiversified Corporate Services Int'l U45118 STR Failure to List Permit Number Guilty 2/25/19 Molony 60 And 60 1/2 Cannon St,Company Llc U33971 Improper Disposal of Garbage/Trash Guilty- Residential 8/29/11 Molony 60 Bull St Llc U31469 Improper Disposal of Garbage/Trash Guilty- Residential 8/29/11 Molony 70 Ashe St. Llc C/O StefanieCompany Lynn Huffer U45433 STR Failure to List Permit Number N/A 5/6/19 Molony 70 Ashe Street Llc C/O CompanyCobb Dill And Hammett U45425 STR Failure to List Permit Number N/A 5/6/19 Molony 78 Smith St. Llc C/O HarrisonCompany Malpass U45427 STR Failure to List Permit Number Guilty 3/25/19 Molony A Lkyon Art And Antiques U18167 Fail To Follow Putout Practices Guilty 1/22/04 Molony Aaron's Deli Rep Chad WalkesCompany U31773 False Alarms Guilty 9/14/16 Molony Abbott Harriet Caroline U79107 Loud & Unnecessary Noise Guilty 8/23/10 Molony Abdo David W U32943 Improper Disposal of Garbage/Trash Guilty- Residential 8/29/11 Molony Abdo David W U37109 Public Nuisances Prohibited Guilty 2/11/14 Pending Abkairian Sabina U41995 1st Offense - Failing to wear face coveringGuilty or mask. -

Outstanding Commissioners
Outstanding Soil and Water Conservation District Commissioner Past Recipients The awards program for Outstanding Soil Conservation District Commissioners began in 1952 with sponsorship from Radio Station WMT. 1952 1956 (sponsored by KXEL, Waterloo) 1 - Ralph Wilcox, Correctionville (Woodbury) 1 - Everett Rice, Storm Lake (Buena Vista) 2 - Guy Wooster, Mapleton (Monona) 2 - Harvey Sornson, Audubon (Audubon) 3 - Ralph Long, Mount Ayr (Ringgold) 3 - Ted Turner, Corning (Adams) 4 - J. C. Skow, Wesley (Kossuth) 4 - John Edwards, Humboldt (Humboldt) 5 - Forrest Hill, Dysart (Tama) 5 - Bruce Bennett, West Des Moines (Polk) 6 - P. W. Spilman, Bloomfield (Davis) 6 - Arlo Noble, Seymour (Wayne) 7 - Garland Byrnes, Dorchester (Allamakee) 7 - Ole Embretson, St. Olaf (Clayton) 8 - Arthur Satterlee,Independence (Buchanan) 8 - John Garner, Davenport (Scott) 9 - Roy White, Salem (Henry) 9 - Clifford Streigle, Delta (Keokuk) 1953 (sponsored by KFMY & KVFD, 1957 (sponsored by Honegger's Feed Fort Dodge; SSCC; IASCDC) Company, Inc.) 1 - Lawrence Everly, Lawton (Woodbury) 1 - Lyle Fogelman, Sutherland (O'Brien) 2 - Wilbur Peters, Persia (Harrison) 2 - C. E. Rich, Glidden (Carroll) 3 - Alvin Peterson, Stanton (Montgomery) 3*- Dallas McGrew, Emerson (Mills) 4 - Lloyd Crom, Chapin (Franklin) 4 - Andrew Christensen, Dows (Franklin) 5 - Howard Sears, Malcom (Poweshiek) 5 - J. Merrill Anderson, Newton (Jasper) 6 - Gilbert Fridley, Indianola (Warren) 6 - Vogel Smith, Russell (Lucas) 7 - Leo Herold, Ft. Atkinson (Winneshiek) 7 - J. Fred Ingels, Maynard (Fayette) -
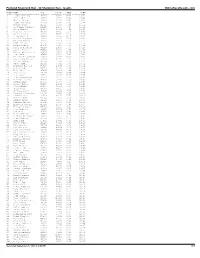
Portland Shamrock Run - 5K Shamrock Run - Results Onlineraceresults.Com
Portland Shamrock Run - 5k Shamrock Run - results OnlineRaceResults.com PLACE NAME DIV DIV PL PACE TIME ----- ---------------------- ------- --------- ------- ----------- 1 Jordan Welling M2529 1/362 4:57 15:22 2 Phil Padilla M2529 2/362 5:04 15:44 3 Ahrlin Bauman M4044 1/404 5:04 15:44 4 Jesse McChesney M2529 3/362 5:05 15:46 5 Robert Cosby M2529 4/362 5:06 15:51 6 Alejandro Cisneros M1519 1/165 5:07 15:51 7 Oscar Bauman M4044 2/404 5:09 16:00 8 Alastair Lawrence M3539 1/425 5:11 16:06 9 Matthew Palmer M2529 5/362 5:14 16:16 10 Ricky Garcia M2024 1/181 5:17 16:23 11 Brian Livingston M3539 2/425 5:26 16:53 12 Matthew Hanson M3034 1/414 5:33 17:13 13 Matt Farley M4549 1/337 5:35 17:18 14 Rich Roberson M3539 3/425 5:35 17:19 15 Cameron Thompson M1519 2/165 5:35 17:20 16 Carrie Dimoff F3034 1/871 5:36 17:23 17 Christopher Meeker M2024 2/181 5:36 17:24 18 Joe Dudman M5054 1/213 5:42 17:41 19 Matthew McCausland M1519 3/165 5:42 17:41 20 Taylor Overmiller M2529 6/362 5:43 17:45 21 Deklan Humble M1519 4/165 5:45 17:51 22 Steve Sample M2529 7/362 5:45 17:51 23 Lorilynn Bloomer F4044 1/681 5:45 17:52 24 Cole Grieb M0114 1/485 5:46 17:53 25 Brian Bernier M3034 2/414 5:46 17:54 26 Ethan Reese M0114 2/485 5:46 17:55 27 Tim Hanson M3539 4/425 5:47 17:56 28 Tim Vanevenhoven M3539 5/425 5:50 18:05 29 Wilfredo Benitez M2024 3/181 5:50 18:05 30 Nathan Hult M4549 2/337 5:50 18:07 31 Ashley Boyle F3034 2/871 5:50 18:08 32 Jason Humble M4044 3/404 5:50 18:08 33 Izaak King M1519 5/165 5:55 18:22 34 Anthony Rinck M4044 4/404 5:57 18:27 35 Makenna Schumacher -

ANTONIS ROKAS, Ph.D. – Brief Curriculum Vitae
ANTONIS ROKAS, Ph.D. – Brief Curriculum Vitae Department of Biological Sciences [email protected] Vanderbilt University office: +1 (615) 936 3892 VU Station B #35-1634, Nashville TN, USA http://www.rokaslab.org/ BRIEF BIOGRAPHY: I am the holder of the Cornelius Vanderbilt Chair in Biological Sciences and Professor in the Departments of Biological Sciences and of Biomedical Informatics at Vanderbilt University. I received my undergraduate degree in Biology from the University of Crete, Greece (1998) and my PhD from Edinburgh University, Scotland (2001). Prior to joining Vanderbilt in the summer of 2007, I was a postdoctoral fellow at the University of Wisconsin-Madison (2002 – 2005) and a research scientist at the Broad Institute (2005 – 2007). Research in my laboratory focuses on the study of the DNA record to gain insight into the patterns and processes of evolution. Through a combination of computational and experimental approaches, my laboratory’s current research aims to understand the molecular foundations of the fungal lifestyle, the reconstruction of the tree of life, and the evolution of human pregnancy. I serve on many journals’ editorial boards, including eLife, Current Biology, BMC Genomics, BMC Microbiology, G3:Genes|Genomes|Genetics, Fungal Genetics & Biology, Evolution, Medicine & Public Health, Frontiers in Microbiology, Microbiology Resource Announcements, and PLoS ONE. My team’s research has been recognized by many awards, including a Searle Scholarship (2008), an NSF CAREER award (2009), a Chancellor’s Award for Research (2011), and an endowed chair (2013). Most recently, I was named a Blavatnik National Awards for Young Scientists Finalist (2017), a Guggenheim Fellow (2018), and a Fellow of the American Academy of Microbiology (2019). -

Beijing 2008 Paralympic Games Cycling (Road) Individual Road Race CP1-2
Die in diesem Dokument zusammengestellten Ergebnislisten haben ihren Ursprung in der IPC-Datenbank http://www.paralympic.org/Athletes/Results und betreffen nur jene Bewerbe, in denen österreichische Athletinnen und Athleten am Start waren. Da diese Listen „Rekonstruktionen“ sind, kann nicht ausgeschlossen werden, dass bei der Übertragung auch Fehler passiert sein können. Sollten Ihnen Fehler auffallen, so können Sie uns diese per E-Mail: [email protected] mitteilen, und wir werden dementsprechende Richtigstellungen veranlassen. Suchfunktion: Diese Datei ist auch mit einer Suchfunktion ausgestattet. Die Suchfunktion ist versionsabhängig und kann dementsprechend unterschiedlich zu handhaben sein. ¾ Bei der Suche können Sie das „Lesezeichen“ verwenden – Lesezeichen bringen Sie schnell zu bestimmten Stellen im Dokument. ¾ Sie können auch direkt das Suchfeld verwenden – bzw. über die Tastenkombination SStrgS+FFF Beijing 2008 Paralympic Games Athletics Men's 100 m T44 Beijing 2008 Paralympic Games Athletics Men's 100 m T44 1st Round Heat 1 Rank Athlete(s) Result 1 Frasure, Brian USA 11.49 2 Shirley, Marlon USA 11.77 3 Wilson, Stephen AUS 11.87 4 Fourie, Arnu RSA 12.05 5 Linhart, Michael AUT 12.11 6 Oliveira, Andre Luiz BRA 12.12 Heat 2 Rank Athlete(s) Result 1 Pistorius, Oscar RSA 11.16 =PR 2 Singleton, Jerome USA 11.48 3 Bausch, Christoph SUI 12.03 4 Marai, Heros ITA 12.05 5 Flores, Antonio MLT 12.71 6 Kim, Vanna CAM 13.45 Final Round Rank Athlete(s) Result 1 Pistorius, Oscar RSA 11.17 2 Singleton, Jerome USA 11.20 3 Frasure, Brian USA 11.50 -

BELGIUM NAME № YEAR Abbeloos, Antoine & Flore (Devos) 944 1975 Abras, Joseph & Jeanne (Delage) 12468 2012 Aelbers, Mathieu H
Righteous Among the Nations Honored by Yad Vashem by 1 January 2020 BELGIUM NAME № YEAR Abbeloos, Antoine & Flore (Devos) 944 1975 Abras, Joseph & Jeanne (Delage) 12468 2012 Aelbers, Mathieu H. & Maria (Alberts) 2794 1984 Aendekerk, Petrus Johannes H. 3109 1985 Aerts Jean 10604 2005 Alardo, Victor & Ida; ch: Marcelle, Marie-Louise, 5483 1992 Joseph Alleman, Jean-Baptiste & Augusta 6978 1996 Amelot, Adele 7357 1996 Anciaux, Emile & Josephine 6466 1994 Andre Emile & daughter Madeleine 8864 2000 Andre Emilie & sister Juliette 8864.1 2000 Andre, Joseph 486 1968 Andre, Paul & Jeanne 2407 1983 Andries, Joseph Jean & Marie (Vanden Bergh) 13158 2015 Andriesse-Renard, Paule 7065 1996 Aneuse, Nelly (Mother Julienne Aneuse) 13034.3 2015 Annaert, Petrus & Isabella, her mother Catherine 11130.1 2007 Steenput-Jacobs Ansiaux, Nicolas J. & Marie Celine 4031.1 1988 Antonis-Maluin, Rene & Leontine (Depraetere) 4435 1989 Arcq, Georgees & Lydie 8432 1999 Arekens, Marie 7465.1 1997 Arnauts, Marie T.E. (Mere Ursule) 11793 2010 Arnoldy, Marcel & Celestine (Schyns) 11350 2008 Arnould, Fernand & Nelly (Bayard) 756 1972 Artus, Mathilde 3127 1985 Audenardt, Pieter & Victorine & daughter Germaine 5986 1994 (Van 't Hoff) Avaux, Evence & Irene (Grimee) 8306 1998 Baeck, Bernard & Arlette (Peeters) 11697 2009 Baeten, Antoon & Maria (Jaspers) 1748 1980 Baggen, Helene 9938 2003 Baichez, Fernand & Juliette-Marie 1638.4 2002 Bal, Auguste & Odile 8998 2000 Bamps, Louis & Elisa 9981 2003 Bams, Joannes & Elvira (Borgugnons) 13402 2017 Bartholomeus, Urbain & Marie-Louise 8910 -
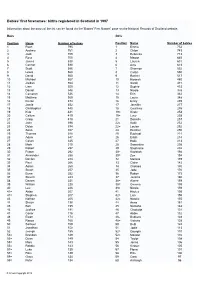
Babies' First Forenames: Births Registered in Scotland in 1997
Babies' first forenames: births registered in Scotland in 1997 Information about the basis of the list can be found via the 'Babies' First Names' page on the National Records of Scotland website. Boys Girls Position Name Number of babies Position Name Number of babies 1 Ryan 795 1 Emma 752 2 Andrew 761 2 Chloe 743 3 Jack 759 3 Rebecca 713 4 Ross 700 4 Megan 645 5 James 638 5 Lauren 631 6 Connor 590 6 Amy 623 7 Scott 586 7 Shannon 552 8 Lewis 568 8 Caitlin 550 9 David 560 9 Rachel 517 10 Michael 557 10 Hannah 480 11 Jordan 554 11 Sarah 471 12 Liam 550 12 Sophie 433 13 Daniel 546 13 Nicole 378 14 Cameron 526 14 Erin 362 15 Matthew 509 15 Laura 348 16 Kieran 474 16 Emily 289 17 Jamie 452 17 Jennifer 277 18 Christopher 440 18 Courtney 276 19 Kyle 421 19= Kirsty 258 20 Callum 419 19= Lucy 258 21 Craig 418 21 Danielle 257 22 John 396 22= Katie 252 23 Dylan 394 22= Louise 252 24 Sean 367 24 Heather 250 25 Thomas 348 25 Rachael 221 26 Adam 347 26 Eilidh 214 27 Calum 335 27 Holly 213 28 Mark 310 28 Samantha 208 29 Robert 297 29 Stephanie 202 30 Fraser 292 30= Kayleigh 194 31 Alexander 288 30= Zoe 194 32 Declan 284 32 Melissa 189 33 Paul 266 33 Claire 182 34 Aaron 260 34 Chelsea 180 35 Stuart 257 35 Jade 176 36 Euan 252 36 Robyn 173 37 Steven 243 37 Jessica 160 38 Darren 231 38= Aimee 159 39 William 228 38= Gemma 159 40 Lee 226 38= Nicola 159 41= Aidan 207 41 Hayley 156 41= Stephen 207 42= Lisa 155 43 Nathan 205 42= Natalie 155 44 Shaun 198 44 Anna 151 45 Ben 195 45 Natasha 148 46 Joshua 191 46 Charlotte 134 47 Conor 176 47 Abbie 132 48 Ewan 174 -

Also in This Issue
Presort Standard U.S. POSTAGE PAID Permit No. 14 Princeton, MN 55371 Iowa Board of Nursing Riverpoint Business Park, 400 SW 8th St., Suite B, Des Moines, IA 50309-4685 Volume 25, Number 3 August, September, October 2006 THE ADVANCED Care is centered on health maintenance and pro- motion. NCSBN SEEKS YOUR INPUT ON REGISTERED NURSE PRACTITIONER ¢ A registered nurse in Iowa may practice at the level of CURRENT LPN/VN The following information is provided to clarify the his/her advanced preparation without registration as an ENTRY-LEVEL NURSING PRACTICE scope of practice, role and functions of the Advanced advanced registered nurse practitioner. Without regis- Registered Nurse Practitioner (ARNP) practicing in Iowa. tration, they may not use the title ARNP, and they do not The National Council of State Boards of Nursing have prescriptive authority. The nurse, employer and ¢ Rules governing the advanced practice aspects of (NCSBN) is about to launch a practice analysis study the physician make the determination regarding policies nursing are found in 655 IAC Chapter 7, which is avail- designed to describe entry-level licensed practical/ and procedures to make prescriptive substances and able on the Iowa Board of Nursing Web site; vocational nurse (LPN/VN) practice. Using a survey devices available to clients. developed by a panel of experts that included both www.state.ia.us/nursing. ¢ Advanced Registered Nurse Practitioner Programs in practicing LPN/VNs from a variety of practice set- ¢ The specialty areas of nursing practice for the ARNP in Iowa: tings and specialty areas as well as LPN/VN educa- Iowa are a certified clinical nurse specialist, certified Allen College – Waterloo tors, a sample of LPN/VNs will be asked to deter- nurse midwife, certified nurse practitioner and a certified Briar Cliff University – Sioux City mine the frequency of performance and the impor- registered nurse anesthetist. -

Provider Directory San Diego Area
Kaiser Permanente for California Medi-Cal Members Provider Directory San Diego Area Kaiser Permanente for California Medi-Cal Members 2001212MC Revised:1/27/2020 Provider Directory This directory provides a list of Kaiser Permanente’s plan providers. To select or change a personal physician or get information about practitioners and services please call 1-888-956-1616 or visit kp.org/finddoctors. Want to know more about your health plan? Need an id card? The Member Service Contact Center is open 24 hours a day, 7 days a week (closed holidays). If you have questions or concerns, we’re here for you. Please call 1-800-464-4000, 1-800-788-0616 (Spanish), 1-800-757-7585 (Chinese dialects), or 711 for TTY users. Some hospitals and other providers do not provide one or more of the following services that may be covered under your plan contract and that you or your family member may need: family planning services, birth control drugs and items, including emergency contraception, surgical birth control, including tubal ligation at the time of labor and delivery, infertility treatments, or abortion. You should get more facts before you enroll. Call your prospective doctor, medical group, independent practice association, or clinic, or call the health plan at 1-800-464-4000, 1-800-788-0616 (Spanish), 1-800-757-7585 (Chinese dialects) to make sure that you can get the services that you need. This directory is for our San Diego service area. Please see the service area description inside for the ZIP codes that are in our service area. -

Origin 1908-1929 Biotech 1930-1984
1914: Operations in Sweden initiated 1926: 1939-40: 1910: 80 200+ 3 employees employees employees Origin 1908-1929 Biotech 1930-1984 1908 1909 1912 1917 1923 1930 1936 1939-40 1945 1946-48 1949 The pharmacists A Albyl® is for In the newly LEO Pharma LEO Pharma Heparin The son-in-law LEO Pharma is the The first bricks From 1949 to Anton Antons bacteriological decades the established enters insulin develops a extraction of Kongsted, first company are laid in 1959, LEO and August and pharmacop most popular laboratories Dr. production in number of method is Knud outside of the UK Ballerup, Pharma moves Kongsted found hysiological painkiller in Marie Krogh Denmark. hormone developed. Abildgaard and the US to Denmark for all production to LEO Pharma in laboratory Denmark. standardises products. takes the reins manufacture LEO Pharma’s the new 1908 when they opens at the digitalis, and the LEO Pharma Hormones are of LEO Pharma. penicillin. new headquarters in receive royal LEO Pharmacy. Albyl® is marketed drug finances the extracted from headquarters. Ballerup, privilege from launched. Digisolvin LEO® development the urine of The LEO Pharma Penicillin, the new Denmark and King Frederik VIII The production is the first and production pregnant price list life-saving drug, is Penicillin begins to run the LEO of sterile and Danish drug to of insulin in the women. The contains considered a production exporting Pharmacy in standardised be exported. 1920s. urine is collected approx. 230 medical miracle. plants are built pharmaceutical Copenhagen. preparations by LEO Pharma branded under licence in products. ® begins in these In 1925, insulin Bottles are staff on products.