A Unified Resource and Configurable Model of the Synapse Proteome And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dynamin Functions and Ligands: Classical Mechanisms Behind
1521-0111/91/2/123–134$25.00 http://dx.doi.org/10.1124/mol.116.105064 MOLECULAR PHARMACOLOGY Mol Pharmacol 91:123–134, February 2017 Copyright ª 2017 by The American Society for Pharmacology and Experimental Therapeutics MINIREVIEW Dynamin Functions and Ligands: Classical Mechanisms Behind Mahaveer Singh, Hemant R. Jadhav, and Tanya Bhatt Department of Pharmacy, Birla Institute of Technology and Sciences Pilani, Pilani Campus, Rajasthan, India Received May 5, 2016; accepted November 17, 2016 Downloaded from ABSTRACT Dynamin is a GTPase that plays a vital role in clathrin-dependent pathophysiology of various disorders, such as Alzheimer’s disease, endocytosis and other vesicular trafficking processes by acting Parkinson’s disease, Huntington’s disease, Charcot-Marie-Tooth as a pair of molecular scissors for newly formed vesicles originating disease, heart failure, schizophrenia, epilepsy, cancer, dominant ’ from the plasma membrane. Dynamins and related proteins are optic atrophy, osteoporosis, and Down s syndrome. This review is molpharm.aspetjournals.org important components for the cleavage of clathrin-coated vesicles, an attempt to illustrate the dynamin-related mechanisms involved phagosomes, and mitochondria. These proteins help in organelle in the above-mentioned disorders and to help medicinal chemists division, viral resistance, and mitochondrial fusion/fission. Dys- to design novel dynamin ligands, which could be useful in the function and mutations in dynamin have been implicated in the treatment of dynamin-related disorders. Introduction GTP hydrolysis–dependent conformational change of GTPase dynamin assists in membrane fission, leading to the generation Dynamins were originally discovered in the brain and identi- of endocytic vesicles (Praefcke and McMahon, 2004; Ferguson at ASPET Journals on September 23, 2021 fied as microtubule binding partners. -

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Chr21 Protein-Protein Interactions: Enrichment in Products Involved in Intellectual Disabilities, Autism and Late Onset Alzheimer Disease
bioRxiv preprint doi: https://doi.org/10.1101/2019.12.11.872606; this version posted December 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Chr21 protein-protein interactions: enrichment in products involved in intellectual disabilities, autism and Late Onset Alzheimer Disease Julia Viard1,2*, Yann Loe-Mie1*, Rachel Daudin1, Malik Khelfaoui1, Christine Plancon2, Anne Boland2, Francisco Tejedor3, Richard L. Huganir4, Eunjoon Kim5, Makoto Kinoshita6, Guofa Liu7, Volker Haucke8, Thomas Moncion9, Eugene Yu10, Valérie Hindie9, Henri Bléhaut11, Clotilde Mircher12, Yann Herault13,14,15,16,17, Jean-François Deleuze2, Jean- Christophe Rain9, Michel Simonneau1, 18, 19, 20** and Aude-Marie Lepagnol- Bestel1** 1 Centre Psychiatrie & Neurosciences, INSERM U894, 75014 Paris, France 2 Laboratoire de génomique fonctionnelle, CNG, CEA, Evry 3 Instituto de Neurociencias CSIC-UMH, Universidad Miguel Hernandez-Campus de San Juan 03550 San Juan (Alicante), Spain 4 Department of Neuroscience, The Johns Hopkins University School of Medicine, Baltimore, MD 21205 USA 5 Center for Synaptic Brain Dysfunctions, Institute for Basic Science, Daejeon 34141, Republic of Korea 6 Department of Molecular Biology, Division of Biological Science, Nagoya University Graduate School of Science, Furo, Chikusa, Nagoya, Japan 7 Department of Biological Sciences, University of Toledo, Toledo, OH, 43606, USA 8 Leibniz Forschungsinstitut für Molekulare Pharmakologie -

Protein Interaction Network of Alternatively Spliced Isoforms from Brain Links Genetic Risk Factors for Autism
ARTICLE Received 24 Aug 2013 | Accepted 14 Mar 2014 | Published 11 Apr 2014 DOI: 10.1038/ncomms4650 OPEN Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism Roser Corominas1,*, Xinping Yang2,3,*, Guan Ning Lin1,*, Shuli Kang1,*, Yun Shen2,3, Lila Ghamsari2,3,w, Martin Broly2,3, Maria Rodriguez2,3, Stanley Tam2,3, Shelly A. Trigg2,3,w, Changyu Fan2,3, Song Yi2,3, Murat Tasan4, Irma Lemmens5, Xingyan Kuang6, Nan Zhao6, Dheeraj Malhotra7, Jacob J. Michaelson7,w, Vladimir Vacic8, Michael A. Calderwood2,3, Frederick P. Roth2,3,4, Jan Tavernier5, Steve Horvath9, Kourosh Salehi-Ashtiani2,3,w, Dmitry Korkin6, Jonathan Sebat7, David E. Hill2,3, Tong Hao2,3, Marc Vidal2,3 & Lilia M. Iakoucheva1 Increased risk for autism spectrum disorders (ASD) is attributed to hundreds of genetic loci. The convergence of ASD variants have been investigated using various approaches, including protein interactions extracted from the published literature. However, these datasets are frequently incomplete, carry biases and are limited to interactions of a single splicing isoform, which may not be expressed in the disease-relevant tissue. Here we introduce a new interactome mapping approach by experimentally identifying interactions between brain-expressed alternatively spliced variants of ASD risk factors. The Autism Spliceform Interaction Network reveals that almost half of the detected interactions and about 30% of the newly identified interacting partners represent contribution from splicing variants, emphasizing the importance of isoform networks. Isoform interactions greatly contribute to establishing direct physical connections between proteins from the de novo autism CNVs. Our findings demonstrate the critical role of spliceform networks for translating genetic knowledge into a better understanding of human diseases. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Sodium Pumps Mediate Activity-Dependent Changes in Mammalian Motor Networks
906 • The Journal of Neuroscience, January 25, 2017 • 37(4):906–921 Systems/Circuits Sodium Pumps Mediate Activity-Dependent Changes in Mammalian Motor Networks X Laurence D. Picton, XFilipe Nascimento, XMatthew J. Broadhead, XKeith T. Sillar, and XGareth B. Miles School of Psychology and Neuroscience, University of St Andrews, St Andrews KY16 9JP, United Kingdom Ubiquitously expressed sodium pumps are best known for maintaining the ionic gradients and resting membrane potential required for generating action potentials. However, activity- and state-dependent changes in pump activity can also influence neuronal firing and regulate rhythmic network output. Here we demonstrate that changes in sodium pump activity regulate locomotor networks in the spinal cord of neonatal mice. The sodium pump inhibitor, ouabain, increased the frequency and decreased the amplitude of drug-induced locomotor bursting, effects that were dependent on the presence of the neuromodulator dopamine. Conversely, activating the pump with the sodium ionophore monensin decreased burst frequency. When more “natural” locomotor output was evoked using dorsal-root stimulation, ouabain increased burst frequency and extended locomotor episode duration, whereas monensin slowed and shortened episodes. Decreasing the time between dorsal-root stimulation, and therefore interepisode interval, also shortened and slowed activity, suggesting that pump activity encodes information about past network output and contributes to feedforward control of subsequent locomotor bouts. Using whole-cell patch-clamp recordings from spinal motoneurons and interneurons, we describe a long-duration (ϳ60 s), activity-dependent, TTX- and ouabain-sensitive, hyperpolarization (ϳ5 mV), which is mediated by spike-dependent increases in pump activity. The duration of this dynamic pump potential is enhanced by dopamine. -

Conserved and Novel Properties of Clathrin-Mediated Endocytosis in Dictyostelium Discoideum" (2012)
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2012 Conserved and Novel Properties of Clathrin- Mediated Endocytosis in Dictyostelium Discoideum Laura Macro Follow this and additional works at: http://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons Recommended Citation Macro, Laura, "Conserved and Novel Properties of Clathrin-Mediated Endocytosis in Dictyostelium Discoideum" (2012). Student Theses and Dissertations. Paper 163. This Thesis is brought to you for free and open access by Digital Commons @ RU. It has been accepted for inclusion in Student Theses and Dissertations by an authorized administrator of Digital Commons @ RU. For more information, please contact [email protected]. CONSERVED AND NOVEL PROPERTIES OF CLATHRIN- MEDIATED ENDOCYTOSIS IN DICTYOSTELIUM DISCOIDEUM A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Laura Macro June 2012 © Copyright by Laura Macro 2012 CONSERVED AND NOVEL PROPERTIES OF CLATHRIN- MEDIATED ENDOCYTOSIS IN DICTYOSTELIUM DISCOIDEUM Laura Macro, Ph.D. The Rockefeller University 2012 The protein clathrin mediates one of the major pathways of endocytosis from the extracellular milieu and plasma membrane. Clathrin functions with a network of interacting accessory proteins, one of which is the adaptor complex AP-2, to co-ordinate vesicle formation. Disruption of genes involved in clathrin-mediated endocytosis causes embryonic lethality in multicellular animals suggesting that clathrin-mediated endocytosis is a fundamental cellular process. However, loss of clathrin-mediated endocytosis genes in single cell eukaryotes, such as S.cerevisiae (yeast), does not cause lethality, suggesting that clathrin may convey specific advantages for multicellularity. -

Downloads/ (Accessed on 17 January 2020)
cells Review Novel Approaches for Identifying the Molecular Background of Schizophrenia Arkadiy K. Golov 1,2,*, Nikolay V. Kondratyev 1 , George P. Kostyuk 3 and Vera E. Golimbet 1 1 Mental Health Research Center, 34 Kashirskoye shosse, 115522 Moscow, Russian; [email protected] (N.V.K.); [email protected] (V.E.G.) 2 Institute of Gene Biology, Russian Academy of Sciences, 34/5 Vavilova Street, 119334 Moscow, Russian 3 Alekseev Psychiatric Clinical Hospital No. 1, 2 Zagorodnoye shosse, 115191 Moscow, Russian; [email protected] * Correspondence: [email protected] Received: 5 November 2019; Accepted: 16 January 2020; Published: 18 January 2020 Abstract: Recent advances in psychiatric genetics have led to the discovery of dozens of genomic loci associated with schizophrenia. However, a gap exists between the detection of genetic associations and understanding the underlying molecular mechanisms. This review describes the basic approaches used in the so-called post-GWAS studies to generate biological interpretation of the existing population genetic data, including both molecular (creation and analysis of knockout animals, exploration of the transcriptional effects of common variants in human brain cells) and computational (fine-mapping of causal variability, gene set enrichment analysis, partitioned heritability analysis) methods. The results of the crucial studies, in which these approaches were used to uncover the molecular and neurobiological basis of the disease, are also reported. Keywords: schizophrenia; GWAS; causal genetic variants; enhancers; brain epigenomics; genome/epigenome editing 1. Introduction Schizophrenia is a severe mental illness that affects between 0.5% and 0.7% of the human population [1]. Both environmental and genetic factors are thought to be involved in its pathogenesis, with genetic factors playing a key role in disease risk, as the heritability of schizophrenia is estimated to be 70–85% [2,3]. -

Epileptic Encephalopathy-Causing Mutations in DNM1 Impair Synaptic Vesicle Endocytosis
Epileptic encephalopathy-causing mutations in DNM1 impair synaptic vesicle endocytosis Ryan S. Dhindsa ABSTRACT Shelton S. Bradrick, PhD Objective: To elucidate the functional consequences of epileptic encephalopathy–causing de novo Xiaodi Yao, PhD mutations in DNM1 (A177P, K206N, G359A), which encodes a large mechanochemical GTPase Erin L. Heinzen, essential for neuronal synaptic vesicle endocytosis. PharmD, PhD Methods: HeLa and COS-7 cells transfected with wild-type and mutant DNM1 constructs were used Slave Petrovski, PhD for transferrin assays, high-content imaging, colocalization studies, Western blotting, and electron Brian J. Krueger, PhD microscopy (EM). EM was also conducted on the brain sections of mice harboring a middle-domain Michael R. Johnson, Dnm1 mutation (Dnm1Ftfl). DPhil Wayne N. Frankel, PhD Results: We demonstrate that the expression of each mutant protein decreased endocytosis activity Steven Petrou, PhD in a dominant-negative manner. One of the G-domain mutations, K206N, decreased protein levels. Rebecca M. Boumil, PhD The G359A mutation, which occurs in the middle domain, disrupted higher-order DNM1 oligomeriza- Dnm1Ftfl David B. Goldstein, PhD tion. EM of mutant DNM1-transfected HeLa cells and of the mouse brain revealed vesicle defects, indicating that the mutations likely interfere with DNM1’s vesicle scission activity. Conclusion: Together, these data suggest that the dysfunction of vesicle scission during synaptic Correspondence to vesicle endocytosis can lead to serious early-onset epilepsies. Neurol Genet 2015;1:e4; doi: Dr. Goldstein: [email protected] 10.1212/01.NXG.0000464295.65736.da GLOSSARY DAPI 5 4’,6-diamidino-2-phenylindole; EDC 5 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide; EM 5 electron microscopy; GFP 5 green fluorescent protein; LGS 5 Lennox-Gastaut syndrome; RFP 5 red fluorescent protein; RIPA 5 radioimmuno- precipitation assay; TBS 5 Tris-buffered saline; WT 5 wild type. -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

Disease-Related Cellular Protein Networks Differentially Affected
www.nature.com/scientificreports OPEN Disease‑related cellular protein networks diferentially afected under diferent EGFR mutations in lung adenocarcinoma Toshihide Nishimura1,8*, Haruhiko Nakamura1,2,8, Ayako Yachie3,8, Takeshi Hase3,8, Kiyonaga Fujii1,8, Hirotaka Koizumi4, Saeko Naruki4, Masayuki Takagi4, Yukiko Matsuoka3, Naoki Furuya5, Harubumi Kato6,7 & Hisashi Saji2 It is unclear how epidermal growth factor receptor EGFR major driver mutations (L858R or Ex19del) afect downstream molecular networks and pathways. This study aimed to provide information on the infuences of these mutations. The study assessed 36 protein expression profles of lung adenocarcinoma (Ex19del, nine; L858R, nine; no Ex19del/L858R, 18). Weighted gene co-expression network analysis together with analysis of variance-based screening identifed 13 co-expressed modules and their eigen proteins. Pathway enrichment analysis for the Ex19del mutation demonstrated involvement of SUMOylation, epithelial and mesenchymal transition, ERK/mitogen- activated protein kinase signalling via phosphorylation and Hippo signalling. Additionally, analysis for the L858R mutation identifed various pathways related to cancer cell survival and death. With regard to the Ex19del mutation, ROCK, RPS6KA1, ARF1, IL2RA and several ErbB pathways were upregulated, whereas AURK and GSKIP were downregulated. With regard to the L858R mutation, RB1, TSC22D3 and DOCK1 were downregulated, whereas various networks, including VEGFA, were moderately upregulated. In all mutation types, CD80/CD86 (B7), MHC, CIITA and IFGN were activated, whereas CD37 and SAFB were inhibited. Costimulatory immune-checkpoint pathways by B7/CD28 were mainly activated, whereas those by PD-1/PD-L1 were inhibited. Our fndings may help identify potential therapeutic targets and develop therapeutic strategies to improve patient outcomes. -
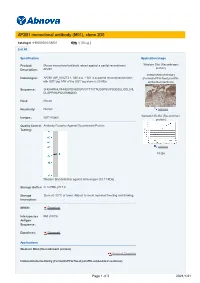
AP2B1 Monoclonal Antibody (M01), Clone 2D5
AP2B1 monoclonal antibody (M01), clone 2D5 Catalog # : H00000163-M01 規格 : [ 50 ug ] List All Specification Application Image Product Mouse monoclonal antibody raised against a partial recombinant Western Blot (Recombinant protein) Description: AP2B1. Immunohistochemistry Immunogen: AP2B1 (NP_001273.1, 585 a.a. ~ 651 a.a) partial recombinant protein (Formalin/PFA-fixed paraffin- with GST tag. MW of the GST tag alone is 26 KDa. embedded sections) Sequence: SHGIHRKHLPIHHGSTDAGDSPVGTTTATNLEQPQVIPSQGDLLGDLLNL DLGPPVNVPQVSSMQMG Host: Mouse Reactivity: Human enlarge Isotype: IgG1 Kappa Sandwich ELISA (Recombinant protein) Quality Control Antibody Reactive Against Recombinant Protein. Testing: enlarge ELISA Western Blot detection against Immunogen (33.11 KDa) . Storage Buffer: In 1x PBS, pH 7.4 Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Instruction: MSDS: Download Interspecies Rat (100%) Antigen Sequence: Datasheet: Download Applications Western Blot (Recombinant protein) Protocol Download Immunohistochemistry (Formalin/PFA-fixed paraffin-embedded sections) Page 1 of 3 2021/1/21 enlarge this image Immunoperoxidase of monoclonal antibody to AP2B1 on formalin-fixed paraffin-embedded human placenta. [antibody concentration 3 ug/ml] Protocol Download Sandwich ELISA (Recombinant protein) Detection limit for recombinant GST tagged AP2B1 is 0.1 ng/ml as a capture antibody. Protocol Download ELISA Gene Information Entrez GeneID: 163 GeneBank NM_001282 Accession#: Protein NP_001273.1 Accession#: Gene Name: AP2B1 Gene Alias: ADTB2,AP105B,AP2-BETA,CLAPB1,DKFZp781K0743 Gene adaptor-related protein complex 2, beta 1 subunit Description: Omim ID: 601025 Gene Ontology: Hyperlink Gene Summary: The protein encoded by this gene is one of two large chain components of the assembly protein complex 2, which serves to link clathrin to receptors in coated vesicles.