Your Genotype Submission Pack
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ocaml Standard Library Ocaml V
OCaml Standard Library OCaml v. 3.12.0 | June 7, 2011 | Copyright c 2011 OCamlPro SAS | http://www.ocamlpro.com/ Standard Modules module List module Array Basic Data Types let len = List.length l let t = Array.create len v let t = Array.init len (fun pos -> v_at_pos) Pervasives All basic functions List.iter (fun ele -> ... ) l; let v = t.(pos) String Functions on Strings let l' = List.map(fun ele -> ... ) l t.(pos) <- v; Array Functions on Polymorphic Arrays let l' = List.rev l1 let len = Array.length t List Functions on Polymorphic Lists let acc' = List.fold left (fun acc ele -> ...) acc l let t' = Array.sub t pos len Char Functions on Characters let acc' = List.fold right (fun ele acc -> ...) l acc let t = Array.of_list list Int32 Functions on 32 bits Integers if List.mem ele l then ... let list = Array.to_list t Int64 Functions on 64 bits Integers if List.for all (fun ele -> ele >= 0) l then ... Array.iter (fun v -> ... ) t; Nativeint Functions on Native Integers if List.exists (fun ele -> ele < 0) l then ... let neg = List.find (fun x -> x < 0) ints Array.iteri (fun pos v -> ... ) t; Advanced Data Types let negs = List.find all (fun x -> x < 0) ints let t' = Array.map (fun v -> ... ) t let t' = Array.mapi (fun pos v -> ... ) t Buffer Automatically resizable strings let (negs,pos) = List.partition (fun x -> x < 0) ints let ele = List.nth 2 list let concat = Array.append prefix suffix Complex Complex Numbers Array.sort compare t; Digest MD5 Checksums let head = List.hd list Hashtbl Polymorphic Hash Tables let tail = List.tl list let value = List.assoc key assocs Queue Polymorphic FIFO module Char Stack Polymorphic LIFO if List.mem assoc key assocs then .. -

High Performance Multi-Node File Copies and Checksums for Clustered File Systems∗
High Performance Multi-Node File Copies and Checksums for Clustered File Systems∗ Paul Z. Kolano, Robert B. Ciotti NASA Advanced Supercomputing Division NASA Ames Research Center, M/S 258-6 Moffett Field, CA 94035 U.S.A. {paul.kolano,bob.ciotti}@nasa.gov Abstract To achieve peak performance from such systems, it is Mcp and msum are drop-in replacements for the stan- typically necessary to utilize multiple concurrent read- dard cp and md5sum programs that utilize multiple types ers/writers from multiple systems to overcome various of parallelism and other optimizations to achieve maxi- single-system limitations such as number of processors mum copy and checksum performance on clustered file and network bandwidth. The standard cp and md5sum systems. Multi-threading is used to ensure that nodes are tools of GNU coreutils [11] found on every modern kept as busy as possible. Read/write parallelism allows Unix/Linux system, however, utilize a single execution individual operations of a single copy to be overlapped thread on a single CPU core of a single system, hence using asynchronous I/O. Multi-node cooperation allows cannot take full advantage of the increased performance different nodes to take part in the same copy/checksum. of clustered file system. Split file processing allows multiple threads to operate This paper describes mcp and msum, which are drop- concurrently on the same file. Finally, hash trees allow in replacements for cp and md5sum that utilize multi- inherently serial checksums to be performed in parallel. ple types of parallelism to achieve maximum copy and This paper presents the design of mcp and msum and de- checksum performance on clustered file systems. -

NOMADS User Guide V1.0
NOMADS User Guide V1.0 Table of Contents • Introduction o Explanation of "Online" and "Offline" Data o Obtaining Offline Data o Offline Order Limitations • Distributed Data Access o Functional Overview o Getting Started with OPeNDAP o Desktop Services ("Clients") o OPeNDAP Servers • Quick Start to Retrieve or Plot Data o Getting Started o Using FTP to Retrieve Data o Using HTTP to Retrieve Data o Using wget to Retrieve Data o Using GDS Clients to Plot Data o Using the NOMADS Server to Create Plots • Advanced Data Access Methods o Subsetting Parameters and Levels Using FTP4u o Obtain ASCII Data Using GDS o Scripting Wget in a Time Loop o Mass GRIB Subsetting: Utilizing Partial-file HTTP Transfers • General Data Structure o Organization of Data Directories o Organization of Data Files o Use of Templates in GrADS Data Descriptor Files • MD5 Hash Files o Purpose of MD5 Files o Identifying MD5 Files o Using md5sum on MD5 Files (Linux) o Using gmd5sum on MD5 Files (FreeBSD) o External Links • Miscellaneous Information o Building Dataset Collections o Working with Timeseries Data o Creating Plots with GrADS o Scheduled Downtime Notification Introduction The model data repository at NCEI contains both deep archived (offline) and online model data. We provide a variety of ways to access our weather and climate model data. You can access the online data using traditional access methods (web-based or FTP), or you can use open and distributed access methods promoted under the collaborative approach called the NOAA National Operational Model Archive and Distribution System (NOMADS). On the Data Products page you are presented with a table that contains basic information about each dataset, as well as links to the various services available for each dataset. -

Download Instructions—Portal
Download instructions These instructions are recommended to download big files. How to download and verify files from downloads.gvsig.org • H ow to download files • G NU/Linux Systems • MacO S X Systems • Windows Systems • H ow to validate the downloaded files How to download files The files distributed on this site can be downloaded using different access protocols, the ones currently available are FTP, HTTP and RSYNC. The base URL of the site for the different protocols is: • ftp://gvsig.org/ • http://downloads.gvsig.org/ • r sync://gvsig.org/downloads/ To download files using the first two protocols is recommended to use client programs able to resume partial downloads, as it is usual to have transfer interruptions when downloading big files like DVD images. There are multiple free (and multi platform) programs to download files using different protocols (in our case we are interested in FTP and HTTP), from them we can highlight curl (http://curl.haxx.se/) and wget (http://www.gnu.org/software/wget/) from the command line ones and Free Download Manager from the GUI ones (this one is only for Windows systems). The curl program is included in MacOS X and is available for almost all GNU/Linux distributions. It can be downloaded in source code or in binary form for different operating systems from the project web site. The wget program is also included in almost all GNU/Linux distributions and its source code or binaries of the program for different systems can be downloaded from this page. Next we will explain how to download files from the most usual operating systems using the programs referenced earlier: • G NU/Linux Systems • MacO S X Systems • Windows Systems The use of rsync (available from the URL http://samba.org/rsync/) it is left as an exercise for the reader, we will only said that it is advised to use the --partial option to avoid problems when there transfers are interrupted. -

High-Performance, Multi-Node File Copies and Checksums for Clustered File Systems Stiffness and Damping Coefficient Estimation O
https://ntrs.nasa.gov/search.jsp?R=20120010444 2019-08-30T20:40:58+00:00Z View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by NASA Technical Reports Server entrance pupil points of interest. These field or aperture, depending on the allo- nates of interest are obtained for every are chosen to be the edge of each space, cations. Once this minimum set of coor- plane of the propagation, the data is for- so that these rays produce the bounding dinates on the pupil and field is ob- matted into an xyz file suitable for FRED volume for the beam. The x and y global tained, a new set of rays is generated optical analysis software to import and coordinate data is collected on the sur- between the field plane and aperture create a STEP file of the data. This re- face planes of interest, typically an image plane (or vice-versa). sults in a spiral-like structure that is easily of the field and entrance pupil internal These rays are then evaluated at imported by mechanical CAD users who of the optical system. This x and y coor- planes between the aperture and field, at can then use an automated algorithm to dinate data is then evaluated using a a desired number of steps perceived nec- wrap a skin around it and create a solid convex hull algorithm, which removes essary to build up the bounding volume that represents the beam. any internal points, which are unneces- or cone shape. At each plane, the ray co- This work was done by Joseph Howard and sary to produce the bounding volume of ordinates are again evaluated using the Lenward Seals of Goddard Space Flight Center. -

HPRC Short Course Introduction to Linux
HPRC Short Course Introduction to Linux Texas A&M University High Performance Research Computing – https://hprc.tamu.edu 1 For Assistance… Website: hprc.tamu.edu Email: [email protected] Telephone: (979) 845-0219 Visit us: Henderson Hall, Room 114A Help us, help you -- we need more info •Which Cluster •NetID •Job ID(s) if any •Location of your job files, input/output files •Application used & module(s) loaded if any •Error messages •Steps you have taken, so we can reproduce the problem Texas A&M University High Performance Research Computing – https://hprc.tamu.edu 2 Course Outline 1 Accessing the System Secure shell 2 Directories Our first commands 3 Gedit An easy text editor 4 File Manipulation Copy, rename/move & remove 5 Passing output & Redirection Learning about operators 6 The Linux file system Attributes and permissions 7 Environment Variables & $PATH Storing and retrieving information 8 Basic Shell Scripting Making Linux work for you Texas A&M University High Performance Research Computing – https://hprc.tamu.edu 3 Your Login Password • Both State of Texas law and TAMU regulations prohibit the sharing and/or illegal use of computer passwords and accounts; • Don’t write down passwords; • Don’t choose easy to guess/crack passwords; • Change passwords frequently Texas A&M University High Performance Research Computing – https://hprc.tamu.edu 4 1. Accessing the system • SSH (secure shell) – The only program allowed for remote access; encrypted communication; freely available for Linux/Unix and Mac OS X hosts; • For Microsoft Windows PCs, use MobaXterm • https://hprc.tamu.edu/wiki/HPRC:MobaXterm – You are able to view images and use GUI applications with MobaXterm – or Putty • https://hprc.tamu.edu/wiki/HPRC:Access#Using_PuTTY – You can not view images or use GUI applications with PuTTY Texas A&M University High Performance Research Computing – https://hprc.tamu.edu 5 Using SSH - MobaXterm on Windows https://hprc.tamu.edu/wiki/HPRC:MobaXterm titan.tamu.edu Use titan.tamu.edu as Remote host name. -
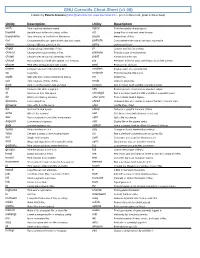
GNU Coreutils Cheat Sheet (V1.00) Created by Peteris Krumins ([email protected], -- Good Coders Code, Great Coders Reuse)
GNU Coreutils Cheat Sheet (v1.00) Created by Peteris Krumins ([email protected], www.catonmat.net -- good coders code, great coders reuse) Utility Description Utility Description arch Print machine hardware name nproc Print the number of processors base64 Base64 encode/decode strings or files od Dump files in octal and other formats basename Strip directory and suffix from file names paste Merge lines of files cat Concatenate files and print on the standard output pathchk Check whether file names are valid or portable chcon Change SELinux context of file pinky Lightweight finger chgrp Change group ownership of files pr Convert text files for printing chmod Change permission modes of files printenv Print all or part of environment chown Change user and group ownership of files printf Format and print data chroot Run command or shell with special root directory ptx Permuted index for GNU, with keywords in their context cksum Print CRC checksum and byte counts pwd Print current directory comm Compare two sorted files line by line readlink Display value of a symbolic link cp Copy files realpath Print the resolved file name csplit Split a file into context-determined pieces rm Delete files cut Remove parts of lines of files rmdir Remove directories date Print or set the system date and time runcon Run command with specified security context dd Convert a file while copying it seq Print sequence of numbers to standard output df Summarize free disk space setuidgid Run a command with the UID and GID of a specified user dir Briefly list directory -

Linux File System and Linux Commands
Hands-on Keyboard: Cyber Experiments for Strategists and Policy Makers Review of the Linux File System and Linux Commands 1. Introduction Becoming adept at using the Linux OS requires gaining familiarity with the Linux file system, file permissions, and a base set of Linux commands. In this activity, you will study how the Linux file system is organized and practice utilizing common Linux commands. Objectives • Describe the purpose of the /bin, /sbin, /etc, /var/log, /home, /proc, /root, /dev, /tmp, and /lib directories. • Describe the purpose of the /etc/shadow and /etc/passwd files. • Utilize a common set of Linux commands including ls, cat, and find. • Understand and manipulate file permissions, including rwx, binary and octal formats. • Change the group and owner of a file. Materials • Windows computer with access to an account with administrative rights The Air Force Cyber College thanks the Advanced Cyber Engineering program at the Air Force Research Laboratory in Rome, NY, for providing the information to assist in educating the general Air Force on the technical aspects of cyberspace. • VirtualBox • Ubuntu OS .iso File Assumptions • The provided instructions were tested on an Ubuntu 15.10 image running on a Windows 8 physical machine. Instructions may vary for other OS. • The student has administrative access to their system and possesses the right to install programs. • The student’s computer has Internet access. 2. Directories / The / directory or root directory is the mother of all Linux directories, containing all of the other directories and files. From a terminal users can type cd/ to move to the root directory. -

Constraints in Dynamic Symbolic Execution: Bitvectors Or Integers?
Constraints in Dynamic Symbolic Execution: Bitvectors or Integers? Timotej Kapus, Martin Nowack, and Cristian Cadar Imperial College London, UK ft.kapus,m.nowack,[email protected] Abstract. Dynamic symbolic execution is a technique that analyses programs by gathering mathematical constraints along execution paths. To achieve bit-level precision, one must use the theory of bitvectors. However, other theories might achieve higher performance, justifying in some cases the possible loss of precision. In this paper, we explore the impact of using the theory of integers on the precision and performance of dynamic symbolic execution of C programs. In particular, we compare an implementation of the symbolic executor KLEE using a partial solver based on the theory of integers, with a standard implementation of KLEE using a solver based on the theory of bitvectors, both employing the popular SMT solver Z3. To our surprise, our evaluation on a synthetic sort benchmark, the ECA set of Test-Comp 2019 benchmarks, and GNU Coreutils revealed that for most applications the integer solver did not lead to any loss of precision, but the overall performance difference was rarely significant. 1 Introduction Dynamic symbolic execution is a popular program analysis technique that aims to systematically explore all the paths in a program. It has been very successful in bug finding and test case generation [3, 4]. The research community and industry have produced many tools performing symbolic execution, such as CREST [5], FuzzBALL [9], KLEE [2], PEX [14], and SAGE [6], among others. To illustrate how dynamic symbolic execution works, consider the program shown in Figure 1a. -

The Linux Command Line
The Linux Command Line Second Internet Edition William E. Shotts, Jr. A LinuxCommand.org Book Copyright ©2008-2013, William E. Shotts, Jr. This work is licensed under the Creative Commons Attribution-Noncommercial-No De- rivative Works 3.0 United States License. To view a copy of this license, visit the link above or send a letter to Creative Commons, 171 Second Street, Suite 300, San Fran- cisco, California, 94105, USA. Linux® is the registered trademark of Linus Torvalds. All other trademarks belong to their respective owners. This book is part of the LinuxCommand.org project, a site for Linux education and advo- cacy devoted to helping users of legacy operating systems migrate into the future. You may contact the LinuxCommand.org project at http://linuxcommand.org. This book is also available in printed form, published by No Starch Press and may be purchased wherever fine books are sold. No Starch Press also offers this book in elec- tronic formats for most popular e-readers: http://nostarch.com/tlcl.htm Release History Version Date Description 13.07 July 6, 2013 Second Internet Edition. 09.12 December 14, 2009 First Internet Edition. 09.11 November 19, 2009 Fourth draft with almost all reviewer feedback incorporated and edited through chapter 37. 09.10 October 3, 2009 Third draft with revised table formatting, partial application of reviewers feedback and edited through chapter 18. 09.08 August 12, 2009 Second draft incorporating the first editing pass. 09.07 July 18, 2009 Completed first draft. Table of Contents Introduction....................................................................................................xvi -

Table of Contents Local Transfers
Table of Contents Local Transfers......................................................................................................1 Checking File Integrity.......................................................................................................1 Local File Transfer Commands...........................................................................................3 Shift Transfer Tool Overview..............................................................................................5 Local Transfers Checking File Integrity It is a good practice to confirm whether your files are complete and accurate before you transfer the files to or from NAS, and again after the transfer is complete. The easiest way to verify the integrity of file transfers is to use the NAS-developed Shift tool for the transfer, with the --verify option enabled. As part of the transfer, Shift will automatically checksum the data at both the source and destination to detect corruption. If corruption is detected, partial file transfers/checksums will be performed until the corruption is rectified. For example: pfe21% shiftc --verify $HOME/filename /nobackuppX/username lou% shiftc --verify /nobackuppX/username/filename $HOME your_localhost% sup shiftc --verify filename pfe: In addition to Shift, there are several algorithms and programs you can use to compute a checksum. If the results of the pre-transfer checksum match the results obtained after the transfer, you can be reasonably certain that the data in the transferred files is not corrupted. If -

Gnu Coreutils Core GNU Utilities for Version 5.93, 2 November 2005
gnu Coreutils Core GNU utilities for version 5.93, 2 November 2005 David MacKenzie et al. This manual documents version 5.93 of the gnu core utilities, including the standard pro- grams for text and file manipulation. Copyright c 1994, 1995, 1996, 2000, 2001, 2002, 2003, 2004, 2005 Free Software Foundation, Inc. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.1 or any later version published by the Free Software Foundation; with no Invariant Sections, with no Front-Cover Texts, and with no Back-Cover Texts. A copy of the license is included in the section entitled “GNU Free Documentation License”. Chapter 1: Introduction 1 1 Introduction This manual is a work in progress: many sections make no attempt to explain basic concepts in a way suitable for novices. Thus, if you are interested, please get involved in improving this manual. The entire gnu community will benefit. The gnu utilities documented here are mostly compatible with the POSIX standard. Please report bugs to [email protected]. Remember to include the version number, machine architecture, input files, and any other information needed to reproduce the bug: your input, what you expected, what you got, and why it is wrong. Diffs are welcome, but please include a description of the problem as well, since this is sometimes difficult to infer. See section “Bugs” in Using and Porting GNU CC. This manual was originally derived from the Unix man pages in the distributions, which were written by David MacKenzie and updated by Jim Meyering.