Density Estimation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nonparametric Density Estimation Using Kernels with Variable Size Windows Anil Ramchandra Londhe Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1980 Nonparametric density estimation using kernels with variable size windows Anil Ramchandra Londhe Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Statistics and Probability Commons Recommended Citation Londhe, Anil Ramchandra, "Nonparametric density estimation using kernels with variable size windows " (1980). Retrospective Theses and Dissertations. 6737. https://lib.dr.iastate.edu/rtd/6737 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This was produced from a copy of a document sent to us for microfilming. WhDe the most advanced technological means to photograph and reproduce this document have been used, the quality is heavily dependent upon the quality of the material submitted. The following explanation of techniques is provided to help you understand markings or notations which may appear on this reproduction. 1. The sign or "target" for pages apparently lacking from the document photographed is "Missing Page(s)". If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting through an image and duplicating adjacent pages to assure you of complete continuity. 2. When an image on the film is obliterated with a round black mark it is an indication that the film inspector noticed either blurred copy because of movement during exposure, or duplicate copy. -

MDL Histogram Density Estimation
MDL Histogram Density Estimation Petri Kontkanen, Petri Myllym¨aki Complex Systems Computation Group (CoSCo) Helsinki Institute for Information Technology (HIIT) University of Helsinki and Helsinki University of Technology P.O.Box 68 (Department of Computer Science) FIN-00014 University of Helsinki, Finland {Firstname}.{Lastname}@hiit.fi Abstract only on finding the optimal bin count. These regu- lar histograms are, however, often problematic. It has been argued (Rissanen, Speed, & Yu, 1992) that reg- We regard histogram density estimation as ular histograms are only good for describing roughly a model selection problem. Our approach uniform data. If the data distribution is strongly non- is based on the information-theoretic min- uniform, the bin count must necessarily be high if one imum description length (MDL) principle, wants to capture the details of the high density portion which can be applied for tasks such as data of the data. This in turn means that an unnecessary clustering, density estimation, image denois- large amount of bins is wasted in the low density re- ing and model selection in general. MDL- gion. based model selection is formalized via the normalized maximum likelihood (NML) dis- To avoid the problems of regular histograms one must tribution, which has several desirable opti- allow the bins to be of variable width. For these irreg- mality properties. We show how this frame- ular histograms, it is necessary to find the optimal set work can be applied for learning generic, ir- of cut points in addition to the number of bins, which regular (variable-width bin) histograms, and naturally makes the learning problem essentially more how to compute the NML model selection difficult. -

A Least-Squares Approach to Direct Importance Estimation∗
JournalofMachineLearningResearch10(2009)1391-1445 Submitted 3/08; Revised 4/09; Published 7/09 A Least-squares Approach to Direct Importance Estimation∗ Takafumi Kanamori [email protected] Department of Computer Science and Mathematical Informatics Nagoya University Furocho, Chikusaku, Nagoya 464-8603, Japan Shohei Hido [email protected] IBM Research Tokyo Research Laboratory 1623-14 Shimotsuruma, Yamato-shi, Kanagawa 242-8502, Japan Masashi Sugiyama [email protected] Department of Computer Science Tokyo Institute of Technology 2-12-1 O-okayama, Meguro-ku, Tokyo 152-8552, Japan Editor: Bianca Zadrozny Abstract We address the problem of estimating the ratio of two probability density functions, which is often referred to as the importance. The importance values can be used for various succeeding tasks such as covariate shift adaptation or outlier detection. In this paper, we propose a new importance estimation method that has a closed-form solution; the leave-one-out cross-validation score can also be computed analytically. Therefore, the proposed method is computationally highly efficient and simple to implement. We also elucidate theoretical properties of the proposed method such as the convergence rate and approximation error bounds. Numerical experiments show that the proposed method is comparable to the best existing method in accuracy, while it is computationally more efficient than competing approaches. Keywords: importance sampling, covariate shift adaptation, novelty detection, regularization path, leave-one-out cross validation 1. Introduction In the context of importance sampling (Fishman, 1996), the ratio of two probability density func- tions is called the importance. The problem of estimating the importance is attracting a great deal of attention these days since the importance can be used for various succeeding tasks such as covariate shift adaptation or outlier detection. -

Lecture 2: Density Estimation 2.1 Histogram
STAT 535: Statistical Machine Learning Autumn 2019 Lecture 2: Density Estimation Instructor: Yen-Chi Chen Main reference: Section 6 of All of Nonparametric Statistics by Larry Wasserman. A book about the methodologies of density estimation: Multivariate Density Estimation: theory, practice, and visualization by David Scott. A more theoretical book (highly recommend if you want to learn more about the theory): Introduction to Nonparametric Estimation by A.B. Tsybakov. Density estimation is the problem of reconstructing the probability density function using a set of given data points. Namely, we observe X1; ··· ;Xn and we want to recover the underlying probability density function generating our dataset. 2.1 Histogram If the goal is to estimate the PDF, then this problem is called density estimation, which is a central topic in statistical research. Here we will focus on the perhaps simplest approach: histogram. For simplicity, we assume that Xi 2 [0; 1] so p(x) is non-zero only within [0; 1]. We also assume that p(x) is smooth and jp0(x)j ≤ L for all x (i.e. the derivative is bounded). The histogram is to partition the set [0; 1] (this region, the region with non-zero density, is called the support of a density function) into several bins and using the count of the bin as a density estimate. When we have M bins, this yields a partition: 1 1 2 M − 2 M − 1 M − 1 B = 0; ;B = ; ; ··· ;B = ; ;B = ; 1 : 1 M 2 M M M−1 M M M M In such case, then for a given point x 2 B`, the density estimator from the histogram will be n number of observations within B` 1 M X p (x) = × = I(X 2 B ): bM n length of the bin n i ` i=1 The intuition of this density estimator is that the histogram assign equal density value to every points within 1 Pn the bin. -

Density Estimation 36-708
Density Estimation 36-708 1 Introduction Let X1;:::;Xn be a sample from a distribution P with density p. The goal of nonparametric density estimation is to estimate p with as few assumptions about p as possible. We denote the estimator by pb. The estimator will depend on a smoothing parameter h and choosing h carefully is crucial. To emphasize the dependence on h we sometimes write pbh. Density estimation used for: regression, classification, clustering and unsupervised predic- tion. For example, if pb(x; y) is an estimate of p(x; y) then we get the following estimate of the regression function: Z mb (x) = ypb(yjx)dy where pb(yjx) = pb(y; x)=pb(x). For classification, recall that the Bayes rule is h(x) = I(p1(x)π1 > p0(x)π0) where π1 = P(Y = 1), π0 = P(Y = 0), p1(x) = p(xjy = 1) and p0(x) = p(xjy = 0). Inserting sample estimates of π1 and π0, and density estimates for p1 and p0 yields an estimate of the Bayes rule. For clustering, we look for the high density regions, based on an estimate of the density. Many classifiers that you are familiar with can be re-expressed this way. Unsupervised prediction is discussed in Section 9. In this case we want to predict Xn+1 from X1;:::;Xn. Example 1 (Bart Simpson) The top left plot in Figure 1 shows the density 4 1 1 X p(x) = φ(x; 0; 1) + φ(x;(j=2) − 1; 1=10) (1) 2 10 j=0 where φ(x; µ, σ) denotes a Normal density with mean µ and standard deviation σ. -

Nonparametric Estimation of Probability Density Functions of Random Persistence Diagrams
Journal of Machine Learning Research 20 (2019) 1-49 Submitted 9/18; Revised 4/19; Published 7/19 Nonparametric Estimation of Probability Density Functions of Random Persistence Diagrams Vasileios Maroulas [email protected] Department of Mathematics University of Tennessee Knoxville, TN 37996, USA Joshua L Mike [email protected] Computational Mathematics, Science, and Engineering Department Michigan State University East Lansing, MI 48823, USA Christopher Oballe [email protected] Department of Mathematics University of Tennessee Knoxville, TN 37996, USA Editor: Boaz Nadler Abstract Topological data analysis refers to a broad set of techniques that are used to make inferences about the shape of data. A popular topological summary is the persistence diagram. Through the language of random sets, we describe a notion of global probability density function for persistence diagrams that fully characterizes their behavior and in part provides a noise likelihood model. Our approach encapsulates the number of topological features and considers the appearance or disappearance of those near the diagonal in a stable fashion. In particular, the structure of our kernel individually tracks long persistence features, while considering those near the diagonal as a collective unit. The choice to describe short persistence features as a group reduces computation time while simultaneously retaining accuracy. Indeed, we prove that the associated kernel density estimate converges to the true distribution as the number of persistence diagrams increases and the bandwidth shrinks accordingly. We also establish the convergence of the mean absolute deviation estimate, defined according to the bottleneck metric. Lastly, examples of kernel density estimation are presented for typical underlying datasets as well as for virtual electroencephalographic data related to cognition. -

DENSITY ESTIMATION INCLUDING EXAMPLES Hans-Georg Müller
DENSITY ESTIMATION INCLUDING EXAMPLES Hans-Georg M¨ullerand Alexander Petersen Department of Statistics University of California Davis, CA 95616 USA In order to gain information about an underlying continuous distribution given a sample of independent data, one has two major options: • Estimate the distribution and probability density function by assuming a finitely- parameterized model for the data and then estimating the parameters of the model by techniques such as maximum likelihood∗(Parametric approach). • Estimate the probability density function nonparametrically by assuming only that it is \smooth" in some sense or falls into some other, appropriately re- stricted, infinite dimensional class of functions (Nonparametric approach). When aiming to assess basic characteristics of a distribution such as skewness∗, tail behavior, number, location and shape of modes∗, or level sets, obtaining an estimate of the probability density function, i.e., the derivative of the distribution function∗, is often a good approach. A histogram∗ is a simple and ubiquitous form of a density estimate, a basic version of which was used already by the ancient Greeks for pur- poses of warfare in the 5th century BC, as described by the historian Thucydides in his History of the Peloponnesian War. Density estimates provide visualization of the distribution and convey considerably more information than can be gained from look- ing at the empirical distribution function, which is another classical nonparametric device to characterize a distribution. This is because distribution functions are constrained to be 0 and 1 and mono- tone in each argument, thus making fine-grained features hard to detect. Further- more, distribution functions are of very limited utility in the multivariate case, while densities remain well defined. -

Density and Distribution Estimation
Density and Distribution Estimation Nathaniel E. Helwig Assistant Professor of Psychology and Statistics University of Minnesota (Twin Cities) Updated 04-Jan-2017 Nathaniel E. Helwig (U of Minnesota) Density and Distribution Estimation Updated 04-Jan-2017 : Slide 1 Copyright Copyright c 2017 by Nathaniel E. Helwig Nathaniel E. Helwig (U of Minnesota) Density and Distribution Estimation Updated 04-Jan-2017 : Slide 2 Outline of Notes 1) PDFs and CDFs 3) Histogram Estimates Overview Overview Estimation problem Bins & breaks 2) Empirical CDFs 4) Kernel Density Estimation Overview KDE basics Examples Bandwidth selection Nathaniel E. Helwig (U of Minnesota) Density and Distribution Estimation Updated 04-Jan-2017 : Slide 3 PDFs and CDFs PDFs and CDFs Nathaniel E. Helwig (U of Minnesota) Density and Distribution Estimation Updated 04-Jan-2017 : Slide 4 PDFs and CDFs Overview Density Functions Suppose we have some variable X ∼ f (x) where f (x) is the probability density function (pdf) of X. Note that we have two requirements on f (x): f (x) ≥ 0 for all x 2 X , where X is the domain of X R X f (x)dx = 1 Example: normal distribution pdf has the form 2 1 − (x−µ) f (x) = p e 2σ2 σ 2π + which is well-defined for all x; µ 2 R and σ 2 R . Nathaniel E. Helwig (U of Minnesota) Density and Distribution Estimation Updated 04-Jan-2017 : Slide 5 PDFs and CDFs Overview Standard Normal Distribution If X ∼ N(0; 1), then X follows a standard normal distribution: 1 2 f (x) = p e−x =2 (1) 2π 0.4 ) x ( 0.2 f 0.0 −4 −2 0 2 4 x Nathaniel E. -

Sparr: Analyzing Spatial Relative Risk Using Fixed and Adaptive Kernel Density Estimation in R
JSS Journal of Statistical Software March 2011, Volume 39, Issue 1. http://www.jstatsoft.org/ sparr: Analyzing Spatial Relative Risk Using Fixed and Adaptive Kernel Density Estimation in R Tilman M. Davies Martin L. Hazelton Jonathan C. Marshall Massey University Massey University Massey University Abstract The estimation of kernel-smoothed relative risk functions is a useful approach to ex- amining the spatial variation of disease risk. Though there exist several options for per- forming kernel density estimation in statistical software packages, there have been very few contributions to date that have focused on estimation of a relative risk function per se. Use of a variable or adaptive smoothing parameter for estimation of the individual densities has been shown to provide additional benefits in estimating relative risk and specific computational tools for this approach are essentially absent. Furthermore, lit- tle attention has been given to providing methods in available software for any kind of subsequent analysis with respect to an estimated risk function. To facilitate analyses in the field, the R package sparr is introduced, providing the ability to construct both fixed and adaptive kernel-smoothed densities and risk functions, identify statistically signifi- cant fluctuations in an estimated risk function through the use of asymptotic tolerance contours, and visualize these objects in flexible and attractive ways. Keywords: density estimation, variable bandwidth, tolerance contours, geographical epidemi- ology, kernel smoothing. 1. Introduction In epidemiological studies it is often of interest to have an understanding of the dispersion of some disease within a given geographical region. A common objective in such analyses is to determine the way in which the `risk' of contraction of the disease varies over the spatial area in which the data has been collected. -

Density Estimation (D = 1) Discrete Density Estimation (D > 1)
Discrete Density Estimation (d = 1) Discrete Density Estimation (d > 1) CPSC 540: Machine Learning Density Estimation Mark Schmidt University of British Columbia Winter 2020 Discrete Density Estimation (d = 1) Discrete Density Estimation (d > 1) Admin Registration forms: I will sign them at the end of class (need to submit prereq form first). Website/Piazza: https://www.cs.ubc.ca/~schmidtm/Courses/540-W20. https://piazza.com/ubc.ca/winterterm22019/cpsc540. Assignment 1 due tonight. Gradescope submissions posted on Piazza. Prereq form submitted separately on Gradescope. Make sure to use your student number as your \name" in Gradescope. You can use late days to submit next week if needed. Today is the last day to add or drop the course. Discrete Density Estimation (d = 1) Discrete Density Estimation (d > 1) Last Time: Structure Prediction \Classic" machine learning: models p(yi j xi), where yi was a single variable. In 340 we used simple distributions like the Gaussian and sigmoid. Structured prediction: yi could be a vector, protein, image, dependency tree,. This requires defining more-complicated distributions. Before considering p(yi j xi) for complicated yi: We'll first consider just modeling p(xi), without worrying about conditioning. Discrete Density Estimation (d = 1) Discrete Density Estimation (d > 1) Density Estimation The next topic we'll focus on is density estimation: 21 0 0 03 2????3 60 1 0 07 6????7 6 7 6 7 X = 60 0 1 07 X~ = 6????7 6 7 6 7 40 1 0 15 4????5 1 0 1 1 ???? What is probability of [1 0 1 1]? Want to estimate probability of feature vectors xi. -
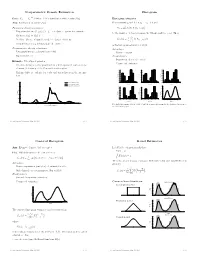
Nonparametric Density Estimation Histogram
Nonparametric Density Estimation Histogram iid Data: X1; : : : ; Xn » P where P is a distribution with density f(x). Histogram estimator Aim: Estimation of density f(x) For constants a0 and h, let ak = a0 + k h and Parametric density estimation: Hk = # Xi Xi 2 (ak¡1; ak] © ¯ ª ± Fit parametric model ff(xj)j 2 £g to data à parameter estimate ^ ¯ be the number of observations in the kth interval (ak¡1; ak]. Then ^ ± Estimate f(x) by f(xj) n ^ 1 f (x) = Hk 1 k¡ k (x) ± Problem: Choice of suitable model à danger of mis¯ts hist hn (a 1;a ] kP=1 ± Complex models (eg mixtures) are di±cult to ¯t is the histogram estimator of f(x). Nonparametric density estimation: Advantages: ± Few assumptions (eg density is smooth) ± Easy to compute ± Exploratory tool Disadvantages: ± Sensitive in choice of o®set a Example: Velocities of galaxies 0 ± Nonsmooth estimator ± Velocities in km/sec of 82 galaxies from 6 well-separated conic sections of an un¯lled survey of the Corona Borealis region. 0.54 0.5 0.56 0.45 0.48 0.4 ± Multimodality is evidence for voids and superclusters in the far uni- 0.40 0.36 0.3 0.32 verse. 0.27 0.24 Density Density 0.2 Density 0.18 0.16 0.1 0.09 0.08 0.00 0.0 0.00 0.25 Kernel estimate (h=0.814) 1 2 3 4 5 6 0.9 1.9 2.9 3.9 4.9 5.9 0.8 1.8 2.8 3.8 4.8 5.8 Kernel estimate (h=0.642) Duration Duration Duration Normal mixture model (k=4) 0.5 0.20 0.56 0.56 0.48 0.48 0.4 0.15 0.40 0.40 0.3 0.32 0.32 Density Density 0.24 Density 0.24 Density 0.2 0.10 0.16 0.16 0.1 0.08 0.08 0.05 0.00 0.00 0.0 0.7 1.7 2.7 3.7 4.7 5.7 0.6 1.6 2.6 3.6 4.6 5.6 1 2 3 4 5 Duration Duration Duration 0.00 0 10 20 30 40 Five shifted histograms with bin width 0.5 and the averaged histogram, for the duration of eruptions of Velocity of galaxy (1000km/s) the Old Faithful geyser. -

APPLIED SMOOTHING TECHNIQUES Part 1: Kernel Density Estimation
APPLIED SMOOTHING TECHNIQUES Part 1: Kernel Density Estimation Walter Zucchini October 2003 Contents 1 Density Estimation 2 1.1 Introduction . 2 1.1.1 The probability density function . 2 1.1.2 Non–parametric estimation of f(x) — histograms . 3 1.2 Kernel density estimation . 3 1.2.1 Weighting functions . 3 1.2.2 Kernels . 7 1.2.3 Densities with bounded support . 8 1.3 Properties of kernel estimators . 12 1.3.1 Quantifying the accuracy of kernel estimators . 12 1.3.2 The bias, variance and mean squared error of fˆ(x) . 12 1.3.3 Optimal bandwidth . 15 1.3.4 Optimal kernels . 15 1.4 Selection of the bandwidth . 16 1.4.1 Subjective selection . 16 1.4.2 Selection with reference to some given distribution . 16 1.4.3 Cross–validation . 18 1.4.4 ”Plug–in” estimator . 19 1.4.5 Summary and extensions . 19 1 Chapter 1 Density Estimation 1.1 Introduction 1.1.1 The probability density function The probability distribution of a continuous–valued random variable X is conventionally described in terms of its probability density function (pdf), f(x), from which probabilities associated with X can be determined using the relationship Z b P (a · X · b) = f(x)dx : a The objective of many investigations is to estimate f(x) from a sample of observations x1; x2; :::; xn . In what follows we will assume that the observations can be regarded as independent realizations of X. The parametric approach for estimating f(x) is to assume that f(x) is a member of some parametric family of distributions, e.g.