Genetic Testing for Nonsyndromic Hearing Loss and Deafness
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
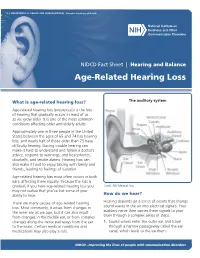
Age-Related Hearing Loss
U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES ∙ National Institutes of Health NIDCD Fact Sheet | Hearing and Balance Age-Related Hearing Loss What is age-related hearing loss? The auditory system Age-related hearing loss (presbycusis) is the loss of hearing that gradually occurs in most of us as we grow older. It is one of the most common conditions affecting older and elderly adults. Approximately one in three people in the United States between the ages of 65 and 74 has hearing loss, and nearly half of those older than 75 have difficulty hearing. Having trouble hearing can make it hard to understand and follow a doctor’s advice, respond to warnings, and hear phones, doorbells, and smoke alarms. Hearing loss can also make it hard to enjoy talking with family and friends, leading to feelings of isolation. Age-related hearing loss most often occurs in both ears, affecting them equally. Because the loss is gradual, if you have age-related hearing loss you Credit: NIH Medical Arts may not realize that you’ve lost some of your ability to hear. How do we hear? There are many causes of age-related hearing Hearing depends on a series of events that change loss. Most commonly, it arises from changes in sound waves in the air into electrical signals. Your the inner ear as we age, but it can also result auditory nerve then carries these signals to your from changes in the middle ear, or from complex brain through a complex series of steps. changes along the nerve pathways from the ear 1. -

Case Study: Testing for Genetic Disorders
Case study: Testing for genetic disorders Purpose: A genetic disorder is a disorder that is caused by an abnormality (or several abnormalities) in a person’s genome. Genetic disorders are often hereditary, which means they are passed down to a child from its parents. The most common genetic disorder is familial hypercholesterolaemia (a genetic predisposition to high cholesterol levels), which is thought to affect 1 in 250 people in the UK. Other disorders, such as cystic fibrosis, are rarer although they can be relatively common in particular ethnic groups; for instance, the incidence of cystic fibrosis in Scotland is almost double the global average. Collectively genetic disorders affect lots of people because there are more than 10,000 different types worldwide. Genetic testing is used to inform people whether they have a disorder, or if there is a chance they might pass a disorder on to their children. Photo credit: jxfzsy/ iStockphoto Photo credit: monkeybusiness/ iStockphoto Approaches to identifying genetic disorders In the UK, if a person is concerned they might have, or might develop, a genetic disorder based on their family history, they can: Go to their GP, who might refer them for genetic testing. Patients usually provide a DNA sample, which is then analysed for a specific abnormality. Usually a single gene will be tested, but whole-genome sequencing approaches are being developed. The 100,000 Genomes Project, for example, aims to study whole-genome sequences from patients with cancer and rare disease Use a commercial genetic testing service. In this case, customers collect samples of their own DNA and send them away for analysis and interpretation Take no action. -

DOI: 10.4274/Jcrpe.Galenos.2021.2020.0175
DOI: 10.4274/jcrpe.galenos.2021.2020.0175 Case report A novel SCNN1A variation in a patient with autosomal-recessive pseudohypoaldosteronism type 1 Mohammed Ayed Huneif1*, Ziyad Hamad AlHazmy2, Anas M. Shoomi 3, Mohammed A. AlGhofely 3, Dr Humariya Heena 5, Aziza M. Mushiba 4, Abdulhamid AlSaheel3 1Pediatric Endocrinologist at Najran university hospital, Najran Saudi Arabia. 2 Pediatric Endocrinologist at Al yamammah hospital, , Riyadh, Saudi Arabia. 3 Pediatric Endocrinologist at Pediatric endocrine department,. Obesity, Endocrine, and Metabolism Center, , King Fahad Medical City, Riyadh, Saudi Arabia. 4Clinical Geneticist, Pediatric Subspecialties Department, Children's Specialized Hospital, King Fahad Medical City, Riyadh, Saudi Arabia. 5 Research Center, King Fahad Medical City, Riyadh , Saudi Arabia What is already known on this topic ? Autosomal-recessive pseudohypoaldosteronism type 1 (PHA1) is a rare genetic disorder caused by different variations in the ENaC subunit genes. Most of these variations appear in SCNN1A mainly in exon eight, which encodes for the alpha subunit of the epithelial sodium channel ENaC. Variations are nonsense, single-base deletions or insertions, or splice site variations, leading to mRNA and proteins of abnormal length. In addition, a few new missense variations have been reported. What this study adds ? We report a novel mutation [ c.729_730delAG (p.Val245Glyfs*65) ] in the exon 4 of the SCNN1A gene In case of autosomal recessive pseudohypoaldosteronism type 1. Patient with PHA1 requires early recognition, proper treatment, and close follow-up. Parents are advised to seek genetic counseling and plan future pregnancies. proof Abstract Pseudohypoaldosteronism type 1 (PHA1) is an autosomal-recessive disorder characterized by defective regulation of body sodium levels. -
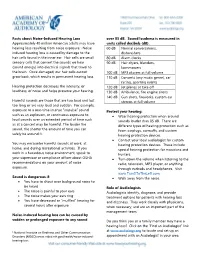
Facts About Noise-Induced Hearing Loss Over 85 Db
Facts about Noise-Induced Hearing Loss over 85 dB. Sound loudness is measured in Approximately 40 million American adults may have units called decibels (dB). hearing loss resulting from noise exposure.1 Noise- 60 dB Normal conversations, induced hearing loss is caused by damage to the dishwashers hair cells found in the inner ear. Hair cells are small 80 dB Alarm clocks sensory cells that convert the sounds we hear 90 dB Hair dryers, blenders, (sound energy) into electrical signals that travel to lawnmowers the brain. Once damaged, our hair cells cannot 100 dB MP3 players at full volume grow back, which results in permanent hearing loss. 110 dB Concerts (any music genre), car racing, sporting events Hearing protection decreases the intensity, or 120 dB Jet planes at take off loudness, of noise and helps preserve your hearing. 130 dB Ambulance, fire engine sirens 140 dB Gun shots, fireworks, custom car Harmful sounds are those that are too loud and last stereos at full volume too long or are very loud and sudden. For example, exposure to a one-time intense “impulse” sound Protect your hearing: such as an explosion, or continuous exposure to • Wear hearing protection when around loud sounds over an extended period of time such sounds louder than 85 dB. There are as at a concert may be harmful. The louder the different types of hearing protection such as sound, the shorter the amount of time you can foam earplugs, earmuffs, and custom safely be around it. hearing protection devices • Contact your local audiologist for custom You may encounter harmful sounds at work, at hearing protection devices. -

Hearing Loss
Randal W. Swenson, M.D. Joshua G. Yorgason, M.D. David K. Palmer, M.D. Wesley R. Brown, M.D. John E. Butler, M.D. Nancy J. Stevenson, PA-C Justin D. Gull, M.D. ENT SPECIALISTS Kristin G. Hoopes, PA-C www.entslc.com Hearing Loss Approximately one in ten persons in the United may result from blockage of the ear canal (wax), States has some degree of hearing loss. Hearing is from a perforation (hole) in the ear drum, or from measured in decibels (dB), and a hearing level of 0- infection or disease of any of the three middle ear 25 dB is considered normal hearing. Your level is: bones. With a conductive loss only, the patient will never go deaf, but will always be able to hear, either Right ear _______ dB Left ear _______dB with reconstructive ear surgery or by use of a properly fitted hearing aid. Some patients who are Hearing Severity / % Loss not candidates for surgery, may benefit from a new 25 dB (normal).….0% 65dB(Severe)……...60% technology, the Baha (bone-anchored hearing aid). 35 dB (mild)……..15% 75dB(Severe)……...75% When there is a problem with the inner ear or 45 dB (moderate)..30% >85dB (Profound)..>90% nerve of hearing, a sensori-neural hearing loss occurs. This is most commonly from normal aging, Normal speech discrimination is 88-100%. Yours is: is usually worse in high frequencies, and can progress to total deafness. Noise exposure is another Right ear _______ % Left ear_______% common cause of high frequency hearing loss. Patients with sensori-neural hearing loss usually complain of difficulty hearing in loud environments. -

Inheriting Genetic Conditions
Help Me Understand Genetics Inheriting Genetic Conditions Reprinted from MedlinePlus Genetics U.S. National Library of Medicine National Institutes of Health Department of Health & Human Services Table of Contents 1 What does it mean if a disorder seems to run in my family? 1 2 Why is it important to know my family health history? 4 3 What are the different ways a genetic condition can be inherited? 6 4 If a genetic disorder runs in my family, what are the chances that my children will have the condition? 15 5 What are reduced penetrance and variable expressivity? 18 6 What do geneticists mean by anticipation? 19 7 What are genomic imprinting and uniparental disomy? 20 8 Are chromosomal disorders inherited? 22 9 Why are some genetic conditions more common in particular ethnic groups? 23 10 What is heritability? 24 Reprinted from MedlinePlus Genetics (https://medlineplus.gov/genetics/) i Inheriting Genetic Conditions 1 What does it mean if a disorder seems to run in my family? A particular disorder might be described as “running in a family” if more than one person in the family has the condition. Some disorders that affect multiple family members are caused by gene variants (also known as mutations), which can be inherited (passed down from parent to child). Other conditions that appear to run in families are not causedby variants in single genes. Instead, environmental factors such as dietary habits, pollutants, or a combination of genetic and environmental factors are responsible for these disorders. It is not always easy to determine whether a condition in a family is inherited. -

Restoration of Fertility by Gonadotropin Replacement in a Man With
J Rohayem and others Fertility in hypogonadotropic 170:4 K11–K17 Case Report CAH with TARTs Restoration of fertility by gonadotropin replacement in a man with hypogonadotropic azoospermia and testicular adrenal rest tumors due to untreated simple virilizing congenital adrenal hyperplasia Julia Rohayem1, Frank Tu¨ ttelmann2, Con Mallidis3, Eberhard Nieschlag1,4, Sabine Kliesch1 and Michael Zitzmann1 Correspondence should be addressed 1Center of Reproductive Medicine and Andrology, Clinical Andrology, University of Muenster, Albert-Schweitzer- to J Rohayem Campus 1, Building D11, D-48149 Muenster, Germany, 2Institute of Human Genetics and 3Institute of Reproductive Email and Regenerative Biology, Center of Reproductive Medicine and Andrology, University of Muenster, Muenster, Julia.Rohayem@ Germany and 4Center of Excellence in Genomic Medicine Research, King Abdulaziz University, Jeddah, Saudi Arabia ukmuenster.de Abstract Context: Classical congenital adrenal hyperplasia (CAH), a genetic disorder characterized by 21-hydroxylase deficiency, impairs male fertility, if insufficiently treated. Patient: A 30-year-old male was referred to our clinic for endocrine and fertility assessment after undergoing unilateral orchiectomy for a suspected testicular tumor. Histopathological evaluation of the removed testis revealed atrophy and testicular adrenal rest tumors (TARTs) and raised the suspicion of underlying CAH. The remaining testis was also atrophic (5 ml) with minor TARTs. Serum 17-hydroxyprogesterone levels were elevated, cortisol levels were at the lower limit of normal range, and gonadotropins at prepubertal levels, but serum testosterone levels were within the normal adult range. Semen analysis revealed azoospermia. CAH was confirmed by a homozygous mutation g.655A/COG (IVS2-13A/COG) in European Journal of Endocrinology CYP21A2. Hydrocortisone (24 mg/m2) administered to suppress ACTH and adrenal androgen overproduction unmasked deficient testicular testosterone production. -

A New Age in the Genetics of Deafness
, September/October 1999. Vol. 1 . No. 6 invited review/cme article A new age in the genetics of deafness Heidi L. Rehtu, MMSC' ot~dCytltllia C. Morton, PAD' Over the last several years, an understanding of the genetics SYNDROMIC AND NONSYNDROMIC DEAFNESS of hearing and deafness has grown exponentially. This progress , The prevalence of severe to profound bilateral congenital has been fueled by fast-paced developments in gene mapping hearing loss is estimated at 1 in 1000 births? When milder I and discovery, leading to the recent cloning and characteriza- forms of permanent hearing impairment at birth are included, tion of many genes critical in the biology of hearing. Among this estimate increases four-fold. Congenital deafness refers to the most significant advances, especially from a clinical per- deafness present at birth, though the cause of such hearing spective, is the discovery that up to 50% of nonsyndromic re- impairment may be genetic (hereditary) or environmental (ac- cessive deafness is caused by mutations in the GIB2 gene, which quired). Various studies estimate that approximately one-half encodes the connexin 26 protein (Cx26).I.' The clinical use of of the cases of congenital deafness are due to genetic factor^,^ screening for mutations in Cx26 and other genes involved in and these factors can be further subdivided by mode of inher- hearing impairment is still a new concept and not widely avail- itance. Approximately 77% of cases of hereditary deafness are able. However, the eventual availability of an array of genetic autosomal recessive, 22% are autosomal dominant, and 1% are tests is likely to have a major impact on the medical decisions X-linked.-l In addition, a small fraction of hereditary deafness made by hearing-impaired patients, their families, and their (< 1%) represents those families with mitochondria1 inheri- physicians. -

Waardenburg Syndrome: Case Series
International Journal of Otorhinolaryngology and Head and Neck Surgery Sachdeva S et al. Int J Otorhinolaryngol Head Neck Surg. 2021 Apr;7(4):668-671 http://www.ijorl.com pISSN 2454-5929 | eISSN 2454-5937 DOI: https://dx.doi.org/10.18203/issn.2454-5929.ijohns20211191 Case Series Waardenburg syndrome: case series Sheenu Sachdeva*, Varunkumar Jayakumar, Shubhlaxmi Atmaram Jaiswal Department of Otorhinolaryngology, Dr. V. M. G. M. C Solapur, Maharashtra, India Received: 13 January 2021 Revised: 16 February 2021 Accepted: 06 March 2021 *Correspondence: Dr. Sheenu Sachdeva, E-mail: [email protected] Copyright: © the author(s), publisher and licensee Medip Academy. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Waardenburg syndrome is a rare genetic disorder of neural crest cell development with incidence of 1:42000 to 1:50,000. The syndrome is not completely expressed and hence adds to its hetergenecity with symptoms varying from one type of syndrome to another and from one patient to another. Unilateral heterochromia that manifests in some people is associated with Waardenburg syndrome and Parry-Romberg syndrome. This is a case series of four cases with features of Waardenburg syndrome with variable presentations and familial inheritance. Keywords: Autosomal dominant deafness, Heterochromia, Pigmentation anomalies, Waardenburg syndrome INTRODUCTION normal. Her mother also had similar hypertelorism with normal hearing (Figure 2). Also there is history suggestive Waardenburg syndrome (WS) is a rare genetic disorder of of premature graying of hair in her grandmother since the neural crest development characterized by varying degrees age of 15 years. -

Preventing Hearing Loss Caused by Chemical (Ototoxicity) and Noise Exposure
Preventing Hearing Loss Caused by Chemical (Ototoxicity) and Noise Exposure Safety and Health Information Bulletin SHIB 03-08-2018 DHHS (NIOSH) Publication No. 2018-124 Introduction Millions of workers are exposed to noise in the workplace every day and when uncontrolled, noise exposure may cause permanent hearing loss. Research demonstrates exposure to certain chemicals, called ototoxicants, may cause hearing loss or balance problems, regardless of noise exposure. Substances including certain pesticides, solvents, and pharmaceuticals that contain ototoxicants can negatively affect how the ear functions, causing hearing loss, and/or affect balance. Source/Copyright: OSHA The risk of hearing loss is increased when workers are exposed to these chemicals while working around elevated noise levels. This combination often results in hearing loss that can be temporary or permanent, depending on the level of noise, the dose of the chemical, and the duration of the exposure. This hearing impairment affects many occupations and industries, from machinists to firefighters. Effects on Hearing Harmful exposure to ototoxicants may occur through inhalation, ingestion, or skin absorption. Health effects caused by ototoxic chemicals vary based on exposure frequency, intensity, duration, workplace exposure to other hazards, and individual factors such as age. Effects may be temporary or permanent, can affect hearing sensitivity and result in a standard threshold shift. Since chemicals can affect central portions of the auditory system (e.g., nerves or nuclei in the central nervous system, the pathways to the brain or in the brain itself), not only do sounds need to be louder to be detected, but also they lose clarity. Specifically, speech discrimination dysfunction, the ability to hear voices separately from background noise, may occur and involve: . -

Hearing Loss in Waardenburg Syndrome: a Systematic Review
Clin Genet 2016: 89: 416–425 © 2015 John Wiley & Sons A/S. Printed in Singapore. All rights reserved Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12631 Review Hearing loss in Waardenburg syndrome: a systematic review Song J., Feng Y., Acke F.R., Coucke P., Vleminckx K., Dhooge I.J. Hearing J. Songa,Y.Fenga, F.R. Ackeb, loss in Waardenburg syndrome: a systematic review. P. Couckec,K.Vleminckxc,d Clin Genet 2016: 89: 416–425. © John Wiley & Sons A/S. Published by and I.J. Dhoogeb John Wiley & Sons Ltd, 2015 aDepartment of Otolaryngology, Xiangya Waardenburg syndrome (WS) is a rare genetic disorder characterized by Hospital, Central South University, Changsha, People’s Republic of China, hearing loss (HL) and pigment disturbances of hair, skin and iris. b Classifications exist based on phenotype and genotype. The auditory Department of Otorhinolaryngology, cDepartment of Medical Genetics, Ghent phenotype is inconsistently reported among the different Waardenburg types University/Ghent University Hospital, and causal genes, urging the need for an up-to-date literature overview on Ghent, Belgium, and dDepartment for this particular topic. We performed a systematic review in search for articles Biomedical Molecular Biology, Ghent describing auditory features in WS patients along with the associated University, Ghent, Belgium genotype. Prevalences of HL were calculated and correlated with the different types and genes of WS. Seventy-three articles were included, describing 417 individual patients. HL was found in 71.0% and was Key words: genotype – hearing loss – predominantly bilateral and sensorineural. Prevalence of HL among the inner ear malformation – phenotype – different clinical types significantly differed (WS1: 52.3%, WS2: 91.6%, Waardenburg syndrome WS3: 57.1%, WS4: 83.5%). -
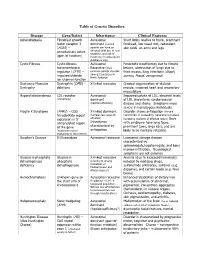
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical Features Achondroplasia Fibroblast growth Autosomal Short limbs relative to trunk, prominent factor receptor 3 dominant (normal forehead, low nasal root, redundant (FGR3) – parents can have an skin folds on arms and legs constitutively active affected child due to new mutation, and risk of (gain of function) recurrence in subsequent children is low) Cystic Fibrosis Cystic fibrosis Autosomal Pancreatic insufficiency due to fibrotic transmembrane Recessive (most lesions, obstruction of lungs due to regulator (CFTR) – common genetic disorder thick mucus, lung infections (Staph, impaired chloride among Caucasians in aureus, Pseud. aeruginosa) North America) ion channel function Duchenne Muscular Dystrophin (DMD) - X-linked recessive Gradual degeneration of skeletal Dystrophy deletions muscle, impaired heart and respiratory musculature Hypercholesterolemia LDL receptor Autosomal Impaired uptake of LDL, elevated levels (commonly) dominant of LDL cholesterol, cardiovascular (haploinsufficiency) disease and stroke. Symptoms more severe in homozygous individuals Fragile X Syndrome (FMR1) – CGG X-linked dominant Disorder shows anticipation (female trinucleotide repeat (females less severely transmitters in succeeding generations produce expansion in 5’ affected) increasing numbers of affected males) Boys untranslated region Inheritance with syndrome have long faces, of the gene characterized by prominent jaws, large ears, and are (expansion occurs anticipation likely to be mentally retarded. exclusively in the mother) Gaucher’s Disease Β-Glucosidase Autosomal recessive Lysosomal storage disease characterized by splenomegaly,hepatomegaly, and bone marrow infiltration. Neurological symptoms are not common Glucose 6-phosphate Glucose 6- X-linked recessive Anemia (due to increased hemolysis) dehydrogenase phosphate (prominent among induced by oxidizing drugs, deficiency dehydrogenase individuals of sulfonamide antibiotics, sulfones (e.g.