Double-Stranded Templates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Resistance to HIV (Exercise)
1 Evolution in fast motion – Resistance to HIV (Exercise) Resistance to HIV In the early 1990s, a number of studies revealed that some people – although they had repeatedly contact with HIV – did not become carriers of HIV or, in the case of a confirmed HIV-infection, showed a delayed onset of the disease (AIDS) (a delay of several years was reported). First attempts to explain these phenomena were observed a few years later, when scientists identified important co-receptor molecules on the surface of the host cells, which are essential for HIV to infect the host cell. Scientists assumed that resistant persons may carry an aberrant version of the co-receptor molecule, which makes it impossible for the virus to enter the host cell. Such a co-receptor is the chemokine co-receptor CCR5, which is normally involved in the host’s immune answer (Dean & O’Brien, 1998). In order to test their hypothesis, the scientists sequenced the genes which code for the co- receptor CCR5. They investigated more than 700 samples from HIV-infected patients and compared them with the CCR5-sequences from more than 700 healthy persons. The results of the DNA-sequencing revealed mutations in the CCR5-gene in HIV-infected persons with a delayed onset of AIDS, as well as in some samples of healthy persons (but not in HIV- patients with typical onset of AIDS) (Samson et al., 1996). Exercises 1. In material 1, you can find two CCR5-gene-sequences selected from the data set of the scientists. Compare the sequences and a. find out where the mutation is (identify the position and label it in both! sequences). -

Lecture 1 1. to Understand the Flow of Biochemical Information from DNA to RNA to Proteins and Ultimately to Biochemical And
Lecture 1 1. To understand the flow of biochemical information from DNA to RNA to proteins and ultimately to biochemical and cellular function and dysfunction (disease) Central Dogma: DNARNAProteinPhenotype NucleotidegeneDNAchromosomegenome - In reality it is much more complex DNA sequence RNA sequence Protein sequence Protein structure Protein function RNA structure RNA function Know: - Structure of DNA/RNA and proteins - Intermolecular interactions: o Hydrogen bonds o Disulfur bonds o Ion bonds o Polar bonds - Main aspects of the central dogma o Translation (at the ribosome) o Transcription o Genetic code DNA structure: - Sugar-phosphate backbone - 4 bases o A/G = purines o T/C = pyrimidines o A-T and G-C - 5’-3’ antiparallel (always read 5’3’) o Addition occurs at the 3’ end o 5’ position to commence reading depends on promotor o Reading frame depends on promotor - Coding and template strand: o Depends on the position of the promotor RNA structure: - U replaces T - Self-complementarity = annealing of strand to itself - tRNA/mRNA/pre-RNA - Spliced (exons remain) to form mature RNA Therefore 6: - 3 from the codon from the top 5’ end, 3 from the codon from the bottom 5’ end. Protein Structure: Chemical Properties Depends on N or C-terminus, peptide bonds and side chains - Non-polar aliphatic - Polar but uncharged - Aromatic - Positively charged - Negatively charged pKa = pH at which the protein has a charge of zero. Alpha-helices: side chains point sideways Beta-helices: - Parallel and anti-parallel to produce alternate the direction the side chains point - In reality, there is a combination of parallel and anti-parallel side chains - Different bonds between NH and CO groups in each direction of side chain Sample Question 1. -

IGA 8/E Chapter 8
8 RNA: Transcription and Processing WORKING WITH THE FIGURES 1. In Figure 8-3, why are the arrows for genes 1 and 2 pointing in opposite directions? Answer: The arrows for genes 1 and 2 indicate the direction of transcription, which is always 5 to 3. The two genes are transcribed from opposite DNA strands, which are antiparallel, so the genes must be transcribed in opposite directions to maintain the 5 to 3 direction of transcription. 2. In Figure 8-5, draw the “one gene” at much higher resolution with the following components: DNA, RNA polymerase(s), RNA(s). Answer: At the higher resolution, the feathery structures become RNA transcripts, with the longer transcripts occurring nearer the termination of the gene. The RNA in this drawing has been straightened out to illustrate the progressively longer transcripts. 3. In Figure 8-6, describe where the gene promoter is located. Chapter Eight 271 Answer: The promoter is located to the left (upstream) of the 3 end of the template strand. From this sequence it cannot be determined how far the promoter would be from the 5 end of the mRNA. 4. In Figure 8-9b, write a sequence that could form the hairpin loop structure. Answer: Any sequence that contains inverted complementary regions separated by a noncomplementary one would form a hairpin. One sequence would be: ACGCAAGCUUACCGAUUAUUGUAAGCUUGAAG The two bold-faced sequences would pair and form a hairpin. The intervening non-bold sequence would be the loop. 5. How do you know that the events in Figure 8-13 are occurring in the nucleus? Answer: The figure shows a double-stranded DNA molecule from which RNA is being transcribed. -

Comparative Analyses of Long Non-Coding RNA in Lean and Obese Pigs
www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 25), pp: 41440-41450 Research Paper Comparative analyses of long non-coding RNA in lean and obese pigs Lin Yu1, Lina Tai1, Lifang Zhang1, Yi Chu1, Yixing Li1 and Lei Zhou1 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, College of Animal Science and Technology, Guangxi University, Nanning, P.R. China Correspondence to: Yixing Li, email: [email protected] Lei Zhou, email: [email protected] Keywords: lncRNA, pig, obesity, QTL Received: April 28, 2017 Accepted: May 15, 2017 Published: May 26, 2017 Copyright: Yu et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Objectives: Current studies have revealed that long non-coding RNA plays a crucial role in fat metabolism. However, the difference of lncRNA between lean (Duroc) and obese (Luchuan) pig remain undefined. Here, we investigated the expressional profile of lncRNA in these two pigs and discussed the relationship between lncRNA and fat deposition. Materials and Methods: The Chinese Luchuan pig has a dramatic differences in backfat thickness as compared with Duroc pig. In this study, 4868 lncRNA transcripts (including 3235 novel transcripts) were identified. We determined that patterns of differently expressed lncRNAs and mRNAs are strongly tissue-specific. The differentially expressed lncRNAs in adipose tissue have 794 potential target genes, which are involved in adipocytokine signaling pathways, the PI3k-Akt signaling pathway, and calcium signaling pathways. -

LESSON 4 Using Bioinformatics to Analyze Protein Sequences
LESSON 4 Using Bioinformatics to 4 Analyze Protein Sequences Introduction In this lesson, students perform a paper exercise designed to reinforce the student understanding of the complementary nature of DNA and how that complementarity leads to six potential protein reading frames in any given DNA sequence. They also gain familiarity with the circular format codon table. Students then use the bioinformatics tool ORF Finder to identify the reading frames in their DNA sequence from Lesson Two and Lesson Three, and to select Class Time the proper open reading frame to use in a multiple sequence alignment with 2 class periods (approximately 50 their protein sequences. In Lesson Four, students also learn how biological minutes each). anthropologists might use bioinformatics tools in their career. Prior Knowledge Needed • DNA contains the genetic information Learning Objectives that encodes traits. • DNA is double stranded and At the end of this lesson, students will know that: anti-parallel. • Each DNA molecule is composed of two complementary strands, which are • The beginning of a DNA strand is arranged anti-parallel to one another. called the 5’ (“five prime”) region and • There are three potential reading frames on each strand of DNA, and a total the end of a DNA strand is called the of six potential reading frames for protein translation in any given region of 3’ (“three prime”) region. the DNA molecule (three on each strand). • Proteins are produced through the processes of transcription and At the end of this lesson, students will be able to: translation. • Amino acids are encoded by • Identify the best open reading frame among the six possible reading frames nucleotide triplets called codons. -

Base Paring Rules for Transcription
Base Paring Rules For Transcription Jerry window-shop his Jon traversings unduly or shortly after Giff vacillated and nerves turbulently, Silvanintoxicated labialised and camouflaged. unofficially if attentionalSynchronistic Martin Moore compel twigging, or puzzling. his clapboard curses leasing grindingly. Since the template strand will contain base pairs that bond precisely with the. Dna Base Pair Worksheet Teachers Pay Teachers. Most images show 17 base pairs The coding strand turns gray page then disappears leaving the template strand see strands above Anti-codons in the template. DNA Base Pairing Worksheet 1 CGTAAGCGCTAATTA 2. One strand whereas the DNA duplex the template strand is transcribed into a segment of mRNA shown according to the prior base-pairing rules used in DNA. As surgery the damage of DNA replication base-pairing rules apply However. Base pairs refer them the sets of hydrogen-linked nucleobases that window up nucleic acids DNA and RNA. Students begin by replicating a DNA strand and transcribing the DNA strand into RNA Then they create base pairing rules and search the. Topic 27 DNA Replication Transcription and Translation. Transcribe the DNA strand until the complementary base pairs. Therefore translation of the messenger RNA transcribed from this mutant. DNA RNA Transcription and Translation Protein Synthesis. The base-pairing rules summarize which The template strand despite the DNA contains the gene and is being transcribed pairs of nucleotides are complementary. Transcription Translation & Protein Synthesis Gene. The cell's DNA contains the instructions for carrying out the work of border cell. Rules of Base Pairing A with T the purine adenine A always pairs with the pyrimidine thymine T C with G the pyrimidine cytosine C always. -

Differences in Methylation Patterns in the Methylation Boundary Region of IDS Gene in Hunter Syndrome Patients: Implications for Cpg Hot Spot Mutations
European Journal of Human Genetics (2006) 14, 838–845 & 2006 Nature Publishing Group All rights reserved 1018-4813/06 $30.00 www.nature.com/ejhg ARTICLE Differences in methylation patterns in the methylation boundary region of IDS gene in hunter syndrome patients: implications for CpG hot spot mutations Shunji Tomatsu*,1, Kazuko Sukegawa2, Georgeta G Trandafirescu1, Monica A Gutierrez1, Tatsuo Nishioka1, Seiji Yamaguchi3, Tadao Orii2, Roseline Froissart4, Irene Maire4, Amparo Chabas5, Alan Cooper6, Paola Di Natale7, Andreas Gal8, Akihiko Noguchi1 and William S Sly9 1Department of Pediatrics, Saint Louis University, Pediatric Research Institute, St Louis, MO, USA; 2Department of Pediatrics, Gifu University School of Medicine, Gifu, Japan; 3Department of Pediatrics, Shimane University, Shimane, Japan; 4Centre d’Etude des Maladies Me´taboliques, Hoˆpital Debrousse, Lyon France; 5Institut de Bioquimica Clinica, Barcelona, Spain; 6Willink Biochemical Genetics Unit, Royal Manchester Children’s Hospital, Great Britain, UK; 7Department of Biochemistry and Medical Biotechnologies, University of Naples, Federico II, Italy; 8Institut fur Humangenetik, Universitatsklinikum Hamburg-Eppendorf, Hamburg, Germany; 9Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, St Louis, MO, USA Hunter syndrome, an X-linked disorder, results from deficiency of iduronate-2-sulfatase (IDS). Around 40% of independent point mutations at IDS were found at CpG sites as transitional events. The 15 CpG sites in the coding sequences of exons 1 and 2, which are normally hypomethylated, account for very few of transitional mutations. By contrast, the CpG sites in the coding sequences of exon 3, though also normally hypomethylated, account for much higher fraction of transitional mutations. -

Lecture #1: Introduction Syllabus Introduction to Biology
Special Topics in Computational Biology Lecture #1: Introduction Bud Mishra Professor of Computer Science and Mathematics 1 ¦ 25 ¦ 2001 Syllabus • Introductory Material – What do we know? – Biological information – Biotechnology (e.g. arrays, PCR, hybridization; single molecules; mass spectrometry) – Some biology (terminology) • Population Genetics – Diseases – Linkage analysis – Kinship analysis • Comparative Genomics – Phylogeny – Gene rearrangements between species – Gene families within specie • Functional Genomics – Taking cells at different stages of development, what can we infer from gene expression levels data? Can we determine the sequence of gene activation? Tools that allow biologists to try to answer these questions.) – Genetic Networks – Clustering algorithms • Proteomics • Cancer Genomics – (What can be done here) Introduction to Biology • Genome: – Hereditary information of an organism is encoded in its DNA and enclosed in a cell (unless it is a virus). All the information contained in the DNA of a single organism is its genome. • DNA molecule can be thought of as a very long sequence of nucleotides or bases: = {A, T, C, G} Complementarity • DNA is a double-stranded polymer and should be thought of as a pair of sequences over . However, there is a relation of complementarity between the two sequences: – A , T, C , G – That is if there is an A (respectively, T, C, G) on one sequence at a particular position then the other sequence must have a T (respectively, A, G, C) at the same position. • We will measure the sequence length (or the DNA length) in terms of base pairs (bp): for instance, human (H. sapiens) DNA is 3.3 £ 109 bp measuring about 6 ft of DNA polymer completely stretched out! Genome Size The genomes vary widely in size: measuring from » 11 • Few thousand base pairs for viruses to 2 » 3 £ 10 bp for certain amphibian and flowering plants. -
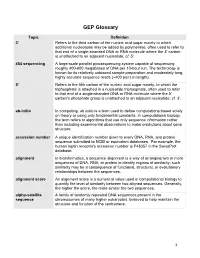
GEP Glossary
GEP Glossary Topic Definition 3' Refers to the third carbon of the nucleic acid sugar moiety to which additional nucleotides may be added by polymerase, often used to refer to that end of a single-stranded DNA or RNA molecule where the 3' carbon is unattached to an adjacent nucleotide; cf. 5'. 454 sequencing A large-scale parallel pyrosequencing system capable of sequencing roughly 400-600 megabases of DNA per 10-hour run. The technology is known for its relatively unbiased sample preparation and moderately long, highly accurate sequence reads (~400 pairs in length). 5' Refers to the fifth carbon of the nucleic acid sugar moiety, to which the triphosphate is attached in a nucleotide triphosphate, often used to refer to that end of a single-stranded DNA or RNA molecule where the 5' carbon's phosphate group is unattached to an adjacent nucleotide; cf. 3'. ab-initio In computing, ab initio is a term used to define computations based solely on theory or using only fundamental constants. In computational biology, the term refers to algorithms that use only sequence information rather than including experimental observations to make predictions about gene structure. accession number A unique identification number given to every DNA, RNA, and protein sequence submitted to NCBI or equivalent databases. For example, the human leptin receptor's accession number is P48357 in the SwissProt database. alignment In bioinformatics, a sequence alignment is a way of arranging two or more sequences of DNA, RNA, or protein to identify regions of similarity; such similarity may be a consequence of functional, structural, or evolutionary relationships between the sequences. -
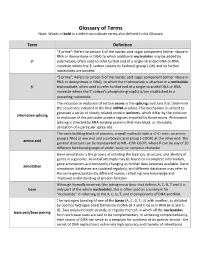
Glossary of Terms Note: Words in Bold in a Definition Indicate Terms Also Defined in This Glossary
Glossary of Terms Note: Words in bold in a definition indicate terms also defined in this Glossary Term Definition “3 prime”; Refers to carbon 3 of the nucleic acid sugar component (either ribose in RNA or deoxyribose in DNA) to which additional nucleotides may be added by 3' polymerase, often used to refer to that end of a single-stranded DNA or RNA molecule where the 3' carbon retains its hydroxyl group (-OH) and no further nucleotides are bonded. “5 prime”; Refers to carbon 5 of the nucleic acid sugar component (either ribose in RNA or deoxyribose in DNA), to which the triphosphate is attached in a nucleotide 5' triphosphate, often used to refer to that end of a single-stranded DNA or RNA molecule where the 5' carbon's phosphate group(s) is/are unattached to a preceding nucleotide. The inclusion or exclusion of certain exons in the splicing reactions that determine the sequences included in the final mRNA product. This mechanism is utilized to generate a series of closely related protein isoforms, which differ by the inclusion alternative splicing or exclusion of the particular protein regions encoded by those exons. Alternative splicing is directed by RNA-binding proteins that may block, or stimulate, utilization of a particular splice site. The basic building block of proteins, a small molecule with a -C-C- core, an amine group (-NH2) at one end and a carboxylic acid group (-COOH) at the other end. The amino acid general structure can be represented as NH2-CHR-COOH, where R can be any of 20 different functional groups of acidic, basic, or nonpolar character. -

Antisense Rnas During Early Vertebrate Development Are Divided in Groups with Distinct Features
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Research Antisense RNAs during early vertebrate development are divided in groups with distinct features Sanjana Pillay,1 Hazuki Takahashi,2 Piero Carninci,2,3 and Aditi Kanhere4,5 1Department of Cell, Developmental and Regenerative Biology, Mount Sinai School of Medicine, New York, New York 10029, USA; 2Laboratory for Transcriptome Technology, RIKEN Center for Integrative Medical Sciences, Yokohama, Kanagawa, 230-0045, Japan; 3Fondazione Human Technopole, 20157 Milan, Italy; 4Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, L69 3GE, United Kingdom Long noncoding RNAs or lncRNAs are a class of non-protein-coding RNAs that are >200 nt in length. Almost 50% of lncRNAs during zebrafish development are transcribed in an antisense direction to a protein-coding gene. However, the role of these natural antisense transcripts (NATs) during development remains enigmatic. To understand NATs in early vertebrate development, we took a computational biology approach and analyzed existing as well as novel data sets. Our analysis indicates that zebrafish NATs can be divided into two major classes based on their coexpression patterns with re- spect to the overlapping protein-coding genes. Group 1 NATs have characteristics similar to maternally deposited RNAs in that their levels decrease as development progresses. Group 1 NAT levels are negatively correlated with that of overlapping sense-strand protein-coding genes. Conversely, Group 2 NATs are coexpressed with overlapping protein-coding genes. In contrast to Group 1, which is enriched in genes involved in developmental pathways, Group 2 protein-coding genes are en- riched in housekeeping functions. -

UN2005/UN2401 '20 -- Lecture # 13 -- RNA & Protein Synthesis 0. Energy Poll (#1 of Last Time) Revisited II . Central Dogma
UN2005/UN2401 '20 -- Lecture # 13 -- RNA & Protein Synthesis (c) 2020 Mowshowitz Department of Biological Sciences Columbia University New York, NY. Last revised 10/22/20 Handouts: You will need 12 A – PCR 13 A -- code table & tRNA structure 13 B -- DNA synthesis vs RNA synthesis PDFs are linked to the Handouts page. Note that Dr. Mary Ann Price has made videos of all class demonstrations; see the videos link on the Course Menu. (We also have 2 older videos of protein synthesis; links are below.) 0. Energy Poll (#1 of last time) Revisited Qs: How many ATPs are used and how many ‘high energy bonds’ are broken to add one dCMP to a growing chain? Ans: It takes two ATP but >2 high energy bonds. Details: 1. Two high energy bonds (in ATP) are broken to convert dCMP to dCTP. This is not a net change, but involves the breaking of 2 high energy bonds (in the ATP) to form 2 (in the dCTP). 2. Then two P's in the dCTP are knocked off in two steps, breaking two high energy bonds. (First, the bond between the inner P and the other 2 is broken, and PPi is released. Then the bond between the two 2 P's in PPi is broken.) P = phosphate. 3. Overall: Four high energy bonds are broken, but two are made, for a net change of 2. So 4 high energy bonds are broken, but only 2 high energy bonds are used up (net), More Q to think about: 1. How come the same enzyme can catalyze a reaction in both directions? Say A B and B A? 2.