Functionless Fraction of the Human Genome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Myth of Junk DNA
The Myth of Junk DNA JoATN h A N W ells s eattle Discovery Institute Press 2011 Description According to a number of leading proponents of Darwin’s theory, “junk DNA”—the non-protein coding portion of DNA—provides decisive evidence for Darwinian evolution and against intelligent design, since an intelligent designer would presumably not have filled our genome with so much garbage. But in this provocative book, biologist Jonathan Wells exposes the claim that most of the genome is little more than junk as an anti-scientific myth that ignores the evidence, impedes research, and is based more on theological speculation than good science. Copyright Notice Copyright © 2011 by Jonathan Wells. All Rights Reserved. Publisher’s Note This book is part of a series published by the Center for Science & Culture at Discovery Institute in Seattle. Previous books include The Deniable Darwin by David Berlinski, In the Beginning and Other Essays on Intelligent Design by Granville Sewell, God and Evolution: Protestants, Catholics, and Jews Explore Darwin’s Challenge to Faith, edited by Jay Richards, and Darwin’s Conservatives: The Misguided Questby John G. West. Library Cataloging Data The Myth of Junk DNA by Jonathan Wells (1942– ) Illustrations by Ray Braun 174 pages, 6 x 9 x 0.4 inches & 0.6 lb, 229 x 152 x 10 mm. & 0.26 kg Library of Congress Control Number: 2011925471 BISAC: SCI029000 SCIENCE / Life Sciences / Genetics & Genomics BISAC: SCI027000 SCIENCE / Life Sciences / Evolution ISBN-13: 978-1-9365990-0-4 (paperback) Publisher Information Discovery Institute Press, 208 Columbia Street, Seattle, WA 98104 Internet: http://www.discoveryinstitutepress.com/ Published in the United States of America on acid-free paper. -

Evolutionary Roles of Transposable Elements – the Science and the Philosophy” University Club, Dalhousie University October 20-21, 2018
“Evolutionary Roles of Transposable Elements – The Science and the Philosophy” University Club, Dalhousie University October 20-21, 2018 ABSTRACTS T. Ryan Gregory (University of Guelph) Junk DNA, genome size, and the onion test It has been known for ~70 years that the amount of DNA in the genomes of eukaryotes varies enormously among species and bears no relation to organismal complexity. Among animals alone, genomes vary more than 7,000-fold in size; across all eukaryotes, the range may exceed 200,000-fold. This extraordinary variability is the result of differential abundances of non-genic DNA, especially in the form of transposable elements. Genome size is correlated with key cellular parameters including cell size and cell division rate, which may lead to organism-level correlations with such features as body size, metabolic rate, and developmental rate. Patterns of genome size variability likely reflect differing evolutionary mechanisms and pressures, from those that add or subtract DNA within genomes to factors that constrain or enhance changes in genome size through organism- level effects. This seminar will provide an overview of the major patterns, correlates, and mechanisms of change in genome size among animals, and will explore the ways in which these are relevant in the debate over potential functions (or lack thereof) for non- genic DNA – as exemplified by the so-called “onion test”. Stefan Linquist (University of Guelph) Four decades debating junk DNA and the Phenotype Paradigm is (somehow) alive and well! A standard trope in molecular biology and genomics says that most types of non-coding DNA have “long been dismissed as junk.” The suggestion is that these regions were neglected by earlier researchers to the extent that hypotheses about their functional significance went untested or were just ignored. -
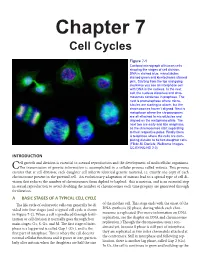
Chapter 7 Cell Cycles
Chapter 7 Cell Cycles Figure 7-1 Confocal micrograph of human cells showing the stages of cell division. DNA is stained blue, microtubules stained green and kinetochores stained pink. Starting from the top and going clockwise you see an interphase cell with DNA in the nucleus. In the next cell, the nucleus dissolves and chro- mosomes condense in prophase. The next is prometaphase where micro- tubules are starting to attach, but the chromosomes haven’t aligned. Next is metaphase where the chromosomes are all attached to microtubules and aligned on the metaphase plate. The next two are early and late anaphase, as the chromosomes start separating to their respective poles. Finally there is telophase where the cells are com- pleting division to be two daughter cells. (Flickr-M. Daniels; Wellcome Images- CC BY-NC-ND 2.0) INTRODUCTION ell growth and division is essential to asexual reproduction and the development of multicellular organisms. CThe transmission of genetic information is accomplished in a cellular process called mitosis. This process ensures that at cell division, each daughter cell inherits identical genetic material, i.e. exactly one copy of each chromosome present in the parental cell. An evolutionary adaptation of mitosis lead to a special type of cell di- vision that reduces the number of chromosomes from diploid to haploid: this is meiosis, and is an essential step in sexual reproduction to avoid doubling the number of chromosomes each time progeny are generated through fertilization. A BASIC STAGES OF A TYPICAL CELL CYCLE The life cycle of eukaryotic cells can generally be di- of the mother cell. -
Junk DNA Neel Jani
JUNK DNA Neel Jani The DNA that composes humans is made of over 3 Types of Junk DNA and their Uses billion base pairs, yet close to 99% of these genes do not code Pseudogenes for proteins and have been termed as “Junk DNA” (Wong et al., 2000), while a more appropriate name would be non- While the preservation of coding DNA (those that do no code for a specific protein), Junk DNA has stirred significant debate and research in the pseudogenes might occur scientific community namely due to its enigmatic nature. Evolutionarily speaking, why should so much energy be because they induce no harm to wasted in the production of something of which only 1% will be functional? Growing research in the past couple of decades the organism, there is growing has thus tried shining light on Junk DNA, namely what it is, BSJ why it exists, its functionality, and its future in humans. evidence suggesting that they History of Junk DNA The concretized term “Junk DNA” originated with regulate prominent diseases such researcher Susumu Ohno in the early 1970’s, yet there has been an unintended misrepresentation from the media that the as diabetes and cancer. discovery of potential functions of noncoding DNA has only begun now. In fact, the discovery of the term went hand-in- When Susumu Ohno coined the term junk DNA, it was in hand with research as to the potential functions for what the reference to the development of pseudogenes. Specifically, term represented. Several researchers dismissed the idea that Ohno described the phenomena of gene duplication; namely, the vast majority of the genome is completely nonfunctional how the cell of an organism could duplicate its genome, and and were receptive to the idea of a function independent of have modifications and alternations to the newly formed copy coding proteins, such as regulation (Gregory and Palazzo, rather than the original. -

“Function” in the Human Genome According to the Evolution-Free Gospel of ENCODE
GBE On the Immortality of Television Sets: “Function” in the Human Genome According to the Evolution-Free Gospel of ENCODE Dan Graur1,*, Yichen Zheng1, Nicholas Price1, Ricardo B.R. Azevedo1, Rebecca A. Zufall1, and Eran Elhaik2 1Department of Biology and Biochemistry, University of Houston 2Department of Mental Health, Johns Hopkins University Bloomberg School of Public Health *Corresponding author: E-mail: [email protected]. Accepted: February 16, 2013 Downloaded from Abstract A recent slew of ENCyclopedia Of DNA Elements (ENCODE) Consortium publications, specifically the article signed by all Consortium members, put forward the idea that more than 80% of the human genome is functional. This claim flies in the face of current http://gbe.oxfordjournals.org/ estimates according to which the fraction of the genome that is evolutionarily conserved through purifying selection is less than 10%. Thus, according to the ENCODE Consortium, a biological function can be maintained indefinitely without selection, which implies that at least 80 À 10 ¼ 70% of the genome is perfectly invulnerable to deleterious mutations, either because no mutation can ever occur in these “functional” regions or because no mutation in these regions can ever be deleterious. This absurd conclusion was reached through various means, chiefly by employing the seldom used “causal role” definition of biological function and then applying it inconsistently to different biochemical properties, by committing a logical fallacy known as “affirming the consequent,” by failing to appreciate the crucial difference between “junk DNA” and “garbage DNA,” by using analytical methods that yield biased errors and inflate estimates of functionality, by favoring statistical sensitivity over specificity, and by emphasizing statistical significance at Carnegie Mellon University on July 3, 2013 rather than the magnitude of the effect. -

Løc Ç to To+Z Lw'le Bound by 1 0285379 .E Bbey Bookblndlng Co
Løc ç to to+z lw'lE Bound by 1 0285379 .E bbey Bookblndlng Co. I ló CathaysTenace, Cardllf Ct2{ {HY Soüth l{â1e3, U.K, Tel: (019) 10395881 ililf I I iltil I lltf I il I ilfilil ilIil UWIC IEARNING CEI'ITRE TIBRARY DIVI$ION.[IÅNDAFF WESTËRI.N AI'ENUE CAROIFF Y cr5 2YË Char acteri s ation of B urkh o I d eri a c ep a ci a from clinical and environmental origins. Paul Wigley BSc. (Hons) University of Wales Institute Cardiff Submiued for the degree of Doctor of Philosophy Discipline : Microbiolo gy September 1998 ABSTRACT Burkholderia cepacia isolates were obtained from the sputum of cystic fìbrosis patients or isolated from the environment. The isolates were characterised phenotypically by determining antibiotic resistance profiles and their ability to macerate onion tissue, and genetically by PCR ribotyping and macro-restriction analysis (genome fingerprinting). The replicon (chromosomal) organisation was determined by pulsed field gel electrophoresis @FGE) and the presence of plasmids was also investigated. Plasmid transfer by conjugation was also investigated. A high degree of genetic and phenotypic variation \ryas found both within and between clinical and environmental populations of B. cepaciø. Environmental isolates were generally less resistant to antibiotics but showed greater ability to macerate onion tissue. Genetic characterisation showed little evidence of acquisition ofB. cepacia from the environment by CF patients, but revealed strong evidence supporting person- to-person transmission ofB. cepacia in the CardiffCF centre and other European centres. Two or more chromosomes were found in 93 Yo of B. cepacla isolates tested, and were also found in other closely related species suggesting such organisation may be common inþ-2 proteobacteria.