Designing Ordered Nucleic Acid Self-Assembly Processes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Not ACID, Not BASE, but SALT a Transaction Processing Perspective on Blockchains
Not ACID, not BASE, but SALT A Transaction Processing Perspective on Blockchains Stefan Tai, Jacob Eberhardt and Markus Klems Information Systems Engineering, Technische Universitat¨ Berlin fst, je, [email protected] Keywords: SALT, blockchain, decentralized, ACID, BASE, transaction processing Abstract: Traditional ACID transactions, typically supported by relational database management systems, emphasize database consistency. BASE provides a model that trades some consistency for availability, and is typically favored by cloud systems and NoSQL data stores. With the increasing popularity of blockchain technology, another alternative to both ACID and BASE is introduced: SALT. In this keynote paper, we present SALT as a model to explain blockchains and their use in application architecture. We take both, a transaction and a transaction processing systems perspective on the SALT model. From a transactions perspective, SALT is about Sequential, Agreed-on, Ledgered, and Tamper-resistant transaction processing. From a systems perspec- tive, SALT is about decentralized transaction processing systems being Symmetric, Admin-free, Ledgered and Time-consensual. We discuss the importance of these dual perspectives, both, when comparing SALT with ACID and BASE, and when engineering blockchain-based applications. We expect the next-generation of decentralized transactional applications to leverage combinations of all three transaction models. 1 INTRODUCTION against. Using the admittedly contrived acronym of SALT, we characterize blockchain-based transactions There is a common belief that blockchains have the – from a transactions perspective – as Sequential, potential to fundamentally disrupt entire industries. Agreed, Ledgered, and Tamper-resistant, and – from Whether we are talking about financial services, the a systems perspective – as Symmetric, Admin-free, sharing economy, the Internet of Things, or future en- Ledgered, and Time-consensual. -

Failures in DBMS
Chapter 11 Database Recovery 1 Failures in DBMS Two common kinds of failures StSystem filfailure (t)(e.g. power outage) ‒ affects all transactions currently in progress but does not physically damage the data (soft crash) Media failures (e.g. Head crash on the disk) ‒ damagg()e to the database (hard crash) ‒ need backup data Recoveryyp scheme responsible for handling failures and restoring database to consistent state 2 Recovery Recovering the database itself Recovery algorithm has two parts ‒ Actions taken during normal operation to ensure system can recover from failure (e.g., backup, log file) ‒ Actions taken after a failure to restore database to consistent state We will discuss (briefly) ‒ Transactions/Transaction recovery ‒ System Recovery 3 Transactions A database is updated by processing transactions that result in changes to one or more records. A user’s program may carry out many operations on the data retrieved from the database, but the DBMS is only concerned with data read/written from/to the database. The DBMS’s abstract view of a user program is a sequence of transactions (reads and writes). To understand database recovery, we must first understand the concept of transaction integrity. 4 Transactions A transaction is considered a logical unit of work ‒ START Statement: BEGIN TRANSACTION ‒ END Statement: COMMIT ‒ Execution errors: ROLLBACK Assume we want to transfer $100 from one bank (A) account to another (B): UPDATE Account_A SET Balance= Balance -100; UPDATE Account_B SET Balance= Balance +100; We want these two operations to appear as a single atomic action 5 Transactions We want these two operations to appear as a single atomic action ‒ To avoid inconsistent states of the database in-between the two updates ‒ And obviously we cannot allow the first UPDATE to be executed and the second not or vice versa. -

SQL Vs Nosql: a Performance Comparison
SQL vs NoSQL: A Performance Comparison Ruihan Wang Zongyan Yang University of Rochester University of Rochester [email protected] [email protected] Abstract 2. ACID Properties and CAP Theorem We always hear some statements like ‘SQL is outdated’, 2.1. ACID Properties ‘This is the world of NoSQL’, ‘SQL is still used a lot by We need to refer the ACID properties[12]: most of companies.’ Which one is accurate? Has NoSQL completely replace SQL? Or is NoSQL just a hype? SQL Atomicity (Structured Query Language) is a standard query language A transaction is an atomic unit of processing; it should for relational database management system. The most popu- either be performed in its entirety or not performed at lar types of RDBMS(Relational Database Management Sys- all. tems) like Oracle, MySQL, SQL Server, uses SQL as their Consistency preservation standard database query language.[3] NoSQL means Not A transaction should be consistency preserving, meaning Only SQL, which is a collection of non-relational data stor- that if it is completely executed from beginning to end age systems. The important character of NoSQL is that it re- without interference from other transactions, it should laxes one or more of the ACID properties for a better perfor- take the database from one consistent state to another. mance in desired fields. Some of the NOSQL databases most Isolation companies using are Cassandra, CouchDB, Hadoop Hbase, A transaction should appear as though it is being exe- MongoDB. In this paper, we’ll outline the general differences cuted in iso- lation from other transactions, even though between the SQL and NoSQL, discuss if Relational Database many transactions are execut- ing concurrently. -

APPENDIX G Acid Dissociation Constants
harxxxxx_App-G.qxd 3/8/10 1:34 PM Page AP11 APPENDIX G Acid Dissociation Constants § ϭ 0.1 M 0 ؍ (Ionic strength ( † ‡ † Name Structure* pKa Ka pKa ϫ Ϫ5 Acetic acid CH3CO2H 4.756 1.75 10 4.56 (ethanoic acid) N ϩ H3 ϫ Ϫ3 Alanine CHCH3 2.344 (CO2H) 4.53 10 2.33 ϫ Ϫ10 9.868 (NH3) 1.36 10 9.71 CO2H ϩ Ϫ5 Aminobenzene NH3 4.601 2.51 ϫ 10 4.64 (aniline) ϪO SNϩ Ϫ4 4-Aminobenzenesulfonic acid 3 H3 3.232 5.86 ϫ 10 3.01 (sulfanilic acid) ϩ NH3 ϫ Ϫ3 2-Aminobenzoic acid 2.08 (CO2H) 8.3 10 2.01 ϫ Ϫ5 (anthranilic acid) 4.96 (NH3) 1.10 10 4.78 CO2H ϩ 2-Aminoethanethiol HSCH2CH2NH3 —— 8.21 (SH) (2-mercaptoethylamine) —— 10.73 (NH3) ϩ ϫ Ϫ10 2-Aminoethanol HOCH2CH2NH3 9.498 3.18 10 9.52 (ethanolamine) O H ϫ Ϫ5 4.70 (NH3) (20°) 2.0 10 4.74 2-Aminophenol Ϫ 9.97 (OH) (20°) 1.05 ϫ 10 10 9.87 ϩ NH3 ϩ ϫ Ϫ10 Ammonia NH4 9.245 5.69 10 9.26 N ϩ H3 N ϩ H2 ϫ Ϫ2 1.823 (CO2H) 1.50 10 2.03 CHCH CH CH NHC ϫ Ϫ9 Arginine 2 2 2 8.991 (NH3) 1.02 10 9.00 NH —— (NH2) —— (12.1) CO2H 2 O Ϫ 2.24 5.8 ϫ 10 3 2.15 Ϫ Arsenic acid HO As OH 6.96 1.10 ϫ 10 7 6.65 Ϫ (hydrogen arsenate) (11.50) 3.2 ϫ 10 12 (11.18) OH ϫ Ϫ10 Arsenious acid As(OH)3 9.29 5.1 10 9.14 (hydrogen arsenite) N ϩ O H3 Asparagine CHCH2CNH2 —— —— 2.16 (CO2H) —— —— 8.73 (NH3) CO2H *Each acid is written in its protonated form. -

Drugs and Acid Dissociation Constants Ionisation of Drug Molecules Most Drugs Ionise in Aqueous Solution.1 They Are Weak Acids Or Weak Bases
Drugs and acid dissociation constants Ionisation of drug molecules Most drugs ionise in aqueous solution.1 They are weak acids or weak bases. Those that are weak acids ionise in water to give acidic solutions while those that are weak bases ionise to give basic solutions. Drug molecules that are weak acids Drug molecules that are weak bases where, HA = acid (the drug molecule) where, B = base (the drug molecule) H2O = base H2O = acid A− = conjugate base (the drug anion) OH− = conjugate base (the drug anion) + + H3O = conjugate acid BH = conjugate acid Acid dissociation constant, Ka For a drug molecule that is a weak acid The equilibrium constant for this ionisation is given by the equation + − where [H3O ], [A ], [HA] and [H2O] are the concentrations at equilibrium. In a dilute solution the concentration of water is to all intents and purposes constant. So the equation is simplified to: where Ka is the acid dissociation constant for the weak acid + + Also, H3O is often written simply as H and the equation for Ka is usually written as: Values for Ka are extremely small and, therefore, pKa values are given (similar to the reason pH is used rather than [H+]. The relationship between pKa and pH is given by the Henderson–Hasselbalch equation: or This relationship is important when determining pKa values from pH measurements. Base dissociation constant, Kb For a drug molecule that is a weak base: 1 Ionisation of drug molecules. 1 Following the same logic as for deriving Ka, base dissociation constant, Kb, is given by: and Ionisation of water Water ionises very slightly. -
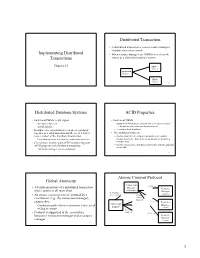
Implementing Distributed Transactions Distributed Transaction Distributed Database Systems ACID Properties Global Atomicity Atom
Distributed Transaction • A distributed transaction accesses resource managers distributed across a network Implementing Distributed • When resource managers are DBMSs we refer to the Transactions system as a distributed database system Chapter 24 DBMS at Site 1 Application Program DBMS 1 at Site 2 2 Distributed Database Systems ACID Properties • Each local DBMS might export • Each local DBMS – stored procedures, or – supports ACID properties locally for each subtransaction – an SQL interface. • Just like any other transaction that executes there • In either case, operations at each site are grouped – eliminates local deadlocks together as a subtransaction and the site is referred • The additional issues are: to as a cohort of the distributed transaction – Global atomicity: all cohorts must abort or all commit – Each subtransaction is treated as a transaction at its site – Global deadlocks: there must be no deadlocks involving • Coordinator module (part of TP monitor) supports multiple sites ACID properties of distributed transaction – Global serialization: distributed transaction must be globally serializable – Transaction manager acts as coordinator 3 4 Atomic Commit Protocol Global Atomicity Transaction (3) xa_reg • All subtransactions of a distributed transaction Manager Resource must commit or all must abort (coordinator) Manager (1) tx_begin (cohort) • An atomic commit protocol, initiated by a (4) tx_commit (5) atomic coordinator (e.g., the transaction manager), commit protocol ensures this. (3) xa_reg Resource Application – Coordinator -

Write-Ahead Logging (WAL) Protocol
CSC 261/461 – Database Systems Lecture 23 Fall 2017 CSC 261, Fall 2017, UR Announcements • Project 3 Due on: 12/01 • Poster: – Will be viewed by the whole department – Even, you may present it later – So, make sure, there is no typo and no embarrassing error. – Go through the poster multiple times – Strongly recommended: • Send us a copy by Sunday. We will try to give you quick feedback. • You can send the poster for printing on Tuesday CSC 261, Fall 2017, UR Today’s Lecture 1. Transactions 2. Properties of Transactions: ACID 3. Logging CSC 261, Fall 2017, UR TRANSACTIONS CSC 261, Fall 2017, UR Transactions: Basic Definition A transaction (“TXN”) is a sequence In the real world, a TXN of one or more operations (reads or either happened writes) which reflects a single real- completely or not at all world transition. START TRANSACTION UPDATE Product SET Price = Price – 1.99 WHERE pname = ‘Gizmo’ COMMIT CSC 261, Fall 2017, UR Transactions: Basic Definition A transaction (“TXN”) is a sequence of one or In the real world, a TXN more operations (reads or writes) which either happened reflects a single real-world transition. completely or not at all Examples: • Transfer money between accounts • Purchase a group of products • Register for a class (either waitlist or allocated) CSC 261, Fall 2017, UR Transactions in SQL • In “ad-hoc” SQL: – Default: each statement = one transaction • In a program, multiple statements can be grouped together as a transaction: START TRANSACTION UPDATE Bank SET amount = amount – 100 WHERE name = ‘Bob’ UPDATE Bank SET amount = amount + 100 WHERE name = ‘Joe’ COMMIT CSC 261, Fall 2017, UR Motivation for Transactions Grouping user actions (reads & writes) into transactions helps with two goals: 1. -

Phosphoric Acid
Common Name: PHOSPHORIC ACID CAS Number: 7664-38-2 RTK Substance number: 1516 DOT Number: UN 1805 Date: May 1997 Revision: April 2004 --------------------------------------------------------------------------- --------------------------------------------------------------------------- HAZARD SUMMARY WORKPLACE EXPOSURE LIMITS * Phosphoric Acid can affect you when breathed in. OSHA: The legal airborne permissible exposure limit * Phosphoric Acid is a CORROSIVE CHEMICAL and (PEL) is 1 mg/m3 averaged over an 8-hour contact can irritate and burn the eyes. workshift. * Breathing Phosphoric Acid can irritate the nose, throat and lungs causing coughing and wheezing. NIOSH: The recommended airborne exposure limit is * Long-term exposure to the liquid may cause drying and 1 mg/m3 averaged over a 10-hour workshift and cracking of the skin. 3 mg/m3, not to be exceeded during any 15 minute work period. IDENTIFICATION Phosphoric Acid is a colorless, odorless solid or a thick, clear ACGIH: The recommended airborne exposure limit is 1 mg/m3 averaged over an 8-hour workshift and liquid. It is used in rustproofing metals, fertilizers, detergents, 3 foods, beverages, and water treatment. 3 mg/m as a STEL (short term exposure limit). REASON FOR CITATION WAYS OF REDUCING EXPOSURE * Phosphoric Acid is on the Hazardous Substance List * Where possible, enclose operations and use local exhaust because it is regulated by OSHA and cited by ACGIH, ventilation at the site of chemical release. If local exhaust DOT, NIOSH, IRIS, NFPA and EPA. ventilation or enclosure is not used, respirators should be * This chemical is on the Special Health Hazard Substance worn. List because it is CORROSIVE. * Wear protective work clothing. * Definitions are provided on page 5. -

Models of Transactions Structuring Applications Flat Transaction Flat
Structuring Applications • Many applications involve long transactions Models of Transactions that make many database accesses • To deal with such complex applications many transaction processing systems Chapter 19 provide mechanisms for imposing some structure on transactions 2 Flat Transaction Flat Transaction • Consists of: begin transaction – Computation on local variables • Abort causes the begin transaction • not seen by DBMS; hence will be ignored in most future discussion execution of a program ¡£ ¤¦©¨ – Access to DBMS using call or ¢¡£ ¥¤§¦©¨ that restores the statement level interface variables updated by the ¡£ ¤¦©¨ • This is transaction schedule; commit ….. applies to these operations ¢¡£ ¤§¦©¨ ….. transaction to the state • No internal structure they had when the if condition then abort • Accesses a single DBMS transaction first accessed commit commit • Adequate for simple applications them. 3 4 Some Limitations of Flat Transactions • Only total rollback (abort) is possible – Partial rollback not possible Providing Structure Within a • All work lost in case of crash Single Transaction • Limited to accessing a single DBMS • Entire transaction takes place at a single point in time 5 6 1 Savepoints begin transaction S1; Savepoints Call to DBMS sp1 := create_savepoint(); S2; sp2 := create_savepoint(); • Problem: Transaction detects condition that S3; requires rollback of recent database changes if (condition) {rollback (sp1); S5}; S4; that it has made commit • Solution 1: Transaction reverses changes • Rollback to spi causes -

Salt: Combining ACID and BASE in a Distributed Database
Salt: Combining ACID and BASE in a Distributed Database Chao Xie, Chunzhi Su, Manos Kapritsos, Yang Wang, Navid Yaghmazadeh, Lorenzo Alvisi, and Prince Mahajan, The University of Texas at Austin https://www.usenix.org/conference/osdi14/technical-sessions/presentation/xie This paper is included in the Proceedings of the 11th USENIX Symposium on Operating Systems Design and Implementation. October 6–8, 2014 • Broomfield, CO 978-1-931971-16-4 Open access to the Proceedings of the 11th USENIX Symposium on Operating Systems Design and Implementation is sponsored by USENIX. Salt: Combining ACID and BASE in a Distributed Database Chao Xie, Chunzhi Su, Manos Kapritsos, Yang Wang, Navid Yaghmazadeh, Lorenzo Alvisi, Prince Mahajan The University of Texas at Austin Abstract: This paper presents Salt, a distributed recently popularized by several NoSQL systems [1, 15, database that allows developers to improve the perfor- 20, 21, 27, 34]. Unlike ACID, BASE offers more of a set mance and scalability of their ACID applications through of programming guidelines (such as the use of parti- the incremental adoption of the BASE approach. Salt’s tion local transactions [32, 37]) than a set of rigorously motivation is rooted in the Pareto principle: for many ap- specified properties and its instantiations take a vari- plications, the transactions that actually test the perfor- ety of application-specific forms. Common among them, mance limits of ACID are few. To leverage this insight, however, is a programming style that avoids distributed Salt introduces BASE transactions, a new abstraction transactions to eliminate the performance and availabil- that encapsulates the workflow of performance-critical ity costs of the associated distributed commit protocol. -

Distributed Systems
Distributed Systems Lec 15: Crashes and Recovery: Write-ahead Logging Slide acks: Dave Andersen (http://www.cs.cmu.edu/~dga/15-440/F10/lectures/Write-ahead-Logging.pdf) Last Few Times (Reminder) • Single-operation consistency – Strict, sequential, causal, and eventual consistency • Multi-operation transactions – ACID properties: atomicity, consistency, isolation, durability • Isolation: two-phase locking (2PL) – Grab locks for all touched objects, then release all locks – Detect or avoid deadlocks by timing out and reverting • Atomicity: two-phase commit (2PC) – Two phases: prepare and commit Two-Phase Commit (Reminder) Transaction Transaction Coordinator (TC) Participant (TP) -- just one -- -- one or more -- TP not allowed to Abort after it’s agreed to Commit Example transfer (X@bank A, Y@bank B, $20) Suppose initially: X.bal = $100 Y.bal = $3 client Bank A Bank B • Clients desire: 1. Atomicity: transfer either happens or not at all 2. Concurrency control: maintain serializability Example transfer (X@bank A, Y@bank B, $20) Suppose initially: X.bal = $100 Y.bal = $3 int transfer(src, dst, amt) { For simplicity, assume the client transaction = begin(); code looks like this: if (src.bal > amt) { int transfer(src, dst, amt) { src.bal -= amt; transaction = begin(); dst.bal += amt; src.bal -= amt; return transaction.commit(); dst.bal += amt; } else { return transaction.commit(); transaction.abort(); } return ABORT; } The banks can unilaterally } decide to COMMIT or ABORT transaction Example TC TP-A TP-B (client or 3rd-party) transaction.commit() prepare B checks if transaction prepare can be committed, if so, blocks rA lock item Y, vote “yes” (use 2PL for this). rB outcome A does similarly (but outcome locks X). -

Dissociation Constants of Organic Acids and Bases
DISSOCIATION CONSTANTS OF ORGANIC ACIDS AND BASES This table lists the dissociation (ionization) constants of over pKa + pKb = pKwater = 14.00 (at 25°C) 1070 organic acids, bases, and amphoteric compounds. All data apply to dilute aqueous solutions and are presented as values of Compounds are listed by molecular formula in Hill order. pKa, which is defined as the negative of the logarithm of the equi- librium constant K for the reaction a References HA H+ + A- 1. Perrin, D. D., Dissociation Constants of Organic Bases in Aqueous i.e., Solution, Butterworths, London, 1965; Supplement, 1972. 2. Serjeant, E. P., and Dempsey, B., Ionization Constants of Organic Acids + - Ka = [H ][A ]/[HA] in Aqueous Solution, Pergamon, Oxford, 1979. 3. Albert, A., “Ionization Constants of Heterocyclic Substances”, in where [H+], etc. represent the concentrations of the respective Katritzky, A. R., Ed., Physical Methods in Heterocyclic Chemistry, - species in mol/L. It follows that pKa = pH + log[HA] – log[A ], so Academic Press, New York, 1963. 4. Sober, H.A., Ed., CRC Handbook of Biochemistry, CRC Press, Boca that a solution with 50% dissociation has pH equal to the pKa of the acid. Raton, FL, 1968. 5. Perrin, D. D., Dempsey, B., and Serjeant, E. P., pK Prediction for Data for bases are presented as pK values for the conjugate acid, a a Organic Acids and Bases, Chapman and Hall, London, 1981. i.e., for the reaction 6. Albert, A., and Serjeant, E. P., The Determination of Ionization + + Constants, Third Edition, Chapman and Hall, London, 1984. BH H + B 7. Budavari, S., Ed., The Merck Index, Twelth Edition, Merck & Co., Whitehouse Station, NJ, 1996.