Using Ancient DNA Techniques to Identify the Origin of Unprovenanced Museum Specimens, As Illustrated by the Identi Cation of a 19Th Century Lion from Amsterdam
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reconstruction of Phylogenetic History to Resolve the Subspecies Anomaly of Pantherine Cats
bioRxiv preprint doi: https://doi.org/10.1101/082891; this version posted October 24, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: Reconstruction of phylogenetic history to resolve the subspecies anomaly of Pantherine cats Authors: Ranajit Das1, Priyanka Upadhyai2 1Manipal Centre for Natural Sciences, Manipal University, Manipal, Karnataka 576104, India 2Department of Medical Genetics, Kasturba Medical College, Manipal University, Manipal, Karnataka 576104, India Email address: [email protected] Running title: Mitogenome phylogeny of Pantherine cats Abstract All charismatic big cats including tiger (Panthera tigris), lion (Panthera leo), leopard (Panthera pardus), snow leopard (Panthera uncial), and jaguar (Panthera onca) are grouped into the subfamily Pantherinae. Several mitogenomic approaches have been employed to reconstruct the phylogenetic history of the Pantherine cats but the phylogeny has remained largely unresolved till date. One of the major reasons for the difficulty in resolving the phylogenetic tree of Pantherine cats is the small sample size. While previous studies included only 5-10 samples, we have used 43 publically available taxa to reconstruct Pantherine phylogenetic history. Complete mtDNA sequences were used from all individuals excluding the control region (15,489bp). A Bayesian MCMC approach was employed to investigate the divergence times among different Pantherine clades. Both maximum likelihood and Bayesian phylogeny generated a dendrogram: Neofelis nebulosa (Panthera tigris (Panthera onca (Panthera uncia (Panthera leo, Panthera pardus)))), grouping lions with leopards and placing snow leopards as an outgroup to this clade. -
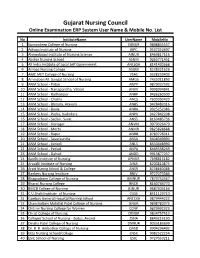
Gujarat Nursing Council Online Examination ERP System User Name & Mobile No
Gujarat Nursing Council Online Examination ERP System User Name & Mobile No. List No InstituteName UserName MobileNo 1 Sumandeep College of Nursing SUNUR 9898855557 2 Adivasi Institute of Nursing AIPC 9537352497 3 Ahmedabad Institute of Nursing Science AINUR 8469817116 4 Akshar Nursing School ASNM 9265771451 5 All India Institute of Local Self Government ANLSGA 8141430568 6 Ambaji Nursing College ASGM 8238321626 7 AMC MET College of Nursing VSAS 9328259403 8 Aminaben M. Gangat School of Nursing AMGS 7435011893 9 ANM School - Patan ANPP 9879037592 10 ANM School - Nanapondha, Valsad ANVV 9998994841 11 ANM School - Radhanpur ANRP 9426260500 12 ANM School - Chotila ANCS 7600050420 13 ANM School - Bhiloda, Aravalli ANBS 9428482016 14 ANM School - Bavla ANBA 9925252386 15 ANM School - Padra, Vadodara ANPV 9427842208 16 ANM School - Sachin, Surat ANSS 8160485736 17 ANM School - Visnagar ANVM 9979326479 18 ANM School - Morbi ANMR 9825828688 19 ANM School - Rapar ANRB 8780726011 20 ANM School - Savarkundla ANSA 9408349990 21 ANM School - Limbdi ANLS 8530448990 22 ANM School - Petlad ANPA 8469538269 23 ANM School - Dahod ANDD 9913877237 24 Apollo Institute of Nursing APNUR 7698815182 25 Aravalli Institute of Nursing AINA 8200810875 26 Arpit Nuring School & College ANSR 8238660088 27 Bankers Nursing Institute BNIV 9727073584 28 Bhagyalaxmi College of Nursing BMNUR 7874752567 29 Bharat Nursing College BNCR 8160744770 30 BMCB College of Nursing BJNUR 9687404164 31 C.U.Shah Institute of Nursing CUSS 8511123710 32 Cambay General Hospital Nursing School ANTCKA 9879444223 33 Chanchalben Mafatlal Patel College of Nursing GNUR 9898780375 34 Chitrini Nursing College for Women CCNP 9829992323 35 Christ College of Nursing CRNUR 9834757510 36 College/ School of Nursing - Zydus, Anand ZSNA 8849216190 37 Dinsha Patel College of Nursing DNNUR 9033183699 38 Dr. -

Population Size and Genetic Diversity of Nigerian Lions (Panthera Leo)
POPULATION SIZE AND GENETIC DIVERSITY OF NIGERIAN LIONS (PANTHERA LEO) POPULATION SIZE AND GENETIC DIVERSITY OF NIGERIAN LIONS (PANTHERA LEO) Talatu Tende AKADEMISK AVHANDLING Som för av filosofie doktorsexamen vid naturvetenskapliga fakulteten, Lunds universitet, kommer att offentligen försvaras i Blå Hallen, Ekologihuset, Lund, Fredagen den 31 January 2014, kl.9:00. ACADEMIC DISSERTATION Presented in fulfilment of the requirements for the degree of Philosophie Doctor at the Faculty of Science, Lund University, to be defended publicly in the Blue Hall, Ecology Building, Sölvegatan 37, Lund, Sweden, Friday 31st January 2014, 9 AM. Fakultetsopponent: Göran Spong, Swedish University of Agricultural Sciences, Umeå POPULATION SIZE AND GENETIC DIVERSITY OF NIGERIAN LIONS (PANTHERA LEO) Talatu Tende A doctoral thesis at a university in Sweden is produced either as a monograph or as a collection of papers. In the latter case, the introductory part constitutes the formal thesis, which summa- rizes the accompanying papers. These have either already been published or are manuscripts at various stages (in press, submitted or in ms). Copyright © Talatu Tende Department of Biology | Lund University Cover art and intro chapter title art: Stina Andersson Layout and formatting: Katarina Eriksson Printed in Sweden at Media Tryck, Lund, 2013 ISBN: 978-91-7473-773-8 CONTENTS List of papers 8 List of contribution 8 Introduction 11 References 30 Popular Summary 37 Acknowledgments 39 Papers Paper I 45 Paper II 57 Paper III 73 Paper IV 85 Paper V 91 7 LIST OF PAPERS This thesis is based on the following papers which are referred to by their Roman numerals. I Ulf Ottosson, Talatu Tende, Christian Hjort & Bengt Hansson. -

North African Lion Fact Sheet
North African Lion Fact Sheet Common Name: North African Lion, Barbary Lion Scientific Name: Panthera leo leo Wild Status: Extinct Habitat: Forests, hills, mountains, plains Country: Egypt, Algeria, Morocco, Libya Shelter: Forests Life Span: Unknown Size: 10ft long Details Present in Roman history and Biblical tales, the Barbary Lion had a reputation as an enormous and vicious creature with a giant mane. Much of their personality and history are, however, exaggerated. This overblown persona made them targets for human hunters, looking to keep their ever expanding territories safe, leading to the extinction of the Barbary Lions. In the wild, they were social mammals who lived in prides, much like the lions of today. They resided in mountainous and hilly areas and often took shelter in forests. Being carnivorous predators, they relied on instinct and teamwork to take down prey such as gazelles. Their fate has often been tied to that of humans who had the ability to catch and control them. The decline of Barbary Lions remains to this day a curious topic for researchers, with efforts being made to locate the purest specimens. Cool Facts • Lions were used as tax payments or lavish gifts. This caused royal families of Morocco to house many Barbary Lions, which eventually made their way to zoos across the world. • These lions are believed to have gone extinct in the 20th century. This would make them one of the most recent extinctions • They are said to have fought gladiators in the Roman empire. The lions present in the Bible are also believed to be Barbary Lions • Many zoos have claimed to have "the last Barbary Lion", however DNA testing has shown these lions are often mixed with other species • Not limited to deserts and savannas, they were often found in forests near mountains • The last Barbary Lion is thought to have been shot in 1942, although some may have survived until the 1960s Taxonomic Breakdown Kingdom: Animalia Phylum: Chordata Class: Mammalia Order: Carnivora Suborder: Feliformia Family: Felidae Subfamily: Pantherinae Genus: Panthera Species: P. -

Husbandry Guidelines for African Lion Panthera Leo Class
Husbandry Guidelines For (Johns 2006) African Lion Panthera leo Class: Mammalia Felidae Compiler: Annemarie Hillermann Date of Preparation: December 2009 Western Sydney Institute of TAFE, Richmond Course Name: Certificate III Captive Animals Course Number: RUV 30204 Lecturer: Graeme Phipps, Jacki Salkeld, Brad Walker DISCLAIMER The information within this document has been compiled by Annemarie Hillermann from general knowledge and referenced sources. This document is strictly for informational purposes only. The information within this document may be amended or changed at any time by the author. The information has been reviewed by professionals within the industry, however, the author will not be held accountable for any misconstrued information within the document. 2 OCCUPATIONAL HEALTH AND SAFETY RISKS Wildlife facilities must adhere to and abide by the policies and procedures of Occupational Health and Safety legislation. A safe and healthy environment must be provided for the animals, visitors and employees at all times within the workplace. All employees must ensure to maintain and be committed to these regulations of OHS within their workplace. All lions are a DANGEROUS/ HIGH RISK and have the potential of fatally injuring a person. Precautions must be followed when working with lions. Consider reducing any potential risks or hazards, including; Exhibit design considerations – e.g. Ergonomics, Chemical, Physical and Mechanical, Behavioural, Psychological, Communications, Radiation, and Biological requirements. EAPA Standards must be followed for exhibit design. Barrier considerations – e.g. Mesh used for roofing area, moats, brick or masonry, Solid/strong metal caging, gates with locking systems, air-locks, double barriers, electric fencing, feeding dispensers/drop slots and ensuring a den area is incorporated. -

Effects of a Combined Enrichment Intervention on the Behavioural and Physiological Welfare Of
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.24.265686; this version posted August 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Effects of a combined enrichment intervention on the behavioural and physiological welfare of 2 captive Asiatic lions (Panthera leo persica) 3 4 Sitendu Goswami1*, Shiv Kumari Patel1, Riyaz Kadivar2, Praveen Chandra Tyagi1, Pradeep 5 Kumar Malik1, Samrat Mondol1* 6 7 1 Wildlife Institute of India, Chandrabani, Dehradun, Uttarakhand, India. 8 2 Sakkarbaug Zoological Garden, Junagadh, Gujarat, India 9 10 11 12 * Corresponding authors: Samrat Mondol, Ph.D., Animal Ecology and Conservation Biology 13 Department, Wildlife Institute of India, Chandrabani, Dehradun, Uttarakhand 248001. Email- 14 [email protected] 15 Sitendu Goswami, Wildlife Institute of India, Chandrabani, Dehradun, Uttarakhand 248001. 16 Email- [email protected] 17 18 19 20 21 22 23 Running head: Impacts of enrichment on Asiatic lions. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.24.265686; this version posted August 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 24 Abstract 25 The endangered Asiatic lion (Panthera leo persica) is currently distributed as a single wild 26 population of 670 individuals and ~400 captive animals globally. -

In Gujarat Sr. Place Dealer Address 1 Amreli Maruti Tractors Opp. Market
In Gujarat Sr. Place Dealer Address 1 Amreli Maruti Tractors Opp. Market Yard, 84, Hirak Baug, Shopping Centre, Amreli 2 Jamkan-Dorna Patel Tractors Agency Nr. Kanya Chhatralaya, Kalawad Road, Jamkandorna 3 Kalavad Gokul Tractors Nr. Shak Market, Nr. Dhoraji Road, Kalavad (Shitla) 4 Keshod Arjun Tractors Mangrol road, Keshod 5 Bhesan Sardar Agency Sardar Patel shopping cente, Nr. S.T. stand, Bhesan, Dist: Junagadh 6 Sadhali Ganesh Engineering Karjan Road, at Sadhali, Dist: Vadodara. Works 7 Talaja Krishna Tractors Ebhal Dwar, Mahuwa Chowkdi, Mahuwa Rd., Talaja 8 Palitana Shriji Tractors Tran Maliya Building, Nr. Madhuli, Bhavanagar Rd., Palitana, Dist: Bhavnagar 9 Bodeli Shri Sainath Motors Laxmi Shopping Centre, Alipura, Chhota Udaipur Rd., Bodeli-391135, Dist: Vadodara 10 Savli Ashok Auto Mobiles Gokul Vatika shopping centre, Opp. New Bus Station, At: Savli, Dist: Vadodara 11 Godhra Hari Om Tractors Old Jakatnaka, Gondhra Nr. Bus Stop, Kalol Rd., Godhra, Panchmahal 12 Bhavnagar Sagar Tractors 303, Trade Cantor, Nr. Bhabniya Blood Bank, Kala Naka, Bhavnagar 13 Chikhli Gayatri Tractor Sales N.H. No. 8, Gandhi Complex, G Floor, Nr. & Service Kalpana Guest House, Chikhli Char Rasta, Chikhli 14 Dhandhuka Rajshakti Tractors Arpan Building, Opp. Birla high school, S.T. Rd., Dhandhuka, Dist: Ahmedabad 15 Patan Brahmani Tractors & G-102, Sidhpur Char Rasta, Patan Jaldhara Irrigation 16 Padra Ashok Automobiles Opp. S.T. Stand, Padra 17 Porbandar Shiv Shakti Sales Jamnagar high way Nr. SBS bank, Porbandar Agency In Other States Sr. Place Dealer Name Address 1 Banglore Sevac Agro India Pvt. Ltd. 217, Kempe Gowda Layout, Ring road, Laggare, Banglore-58, Karnataka 2 Gobichhe- Snusham Farm Machinery & 232, Sathy Main Rd., Opp. -

THE AFRICAN LION (Panthera Leo Leo): a CONTINENT-WIDE SPECIES DISTRIBUTION STUDY and POPULATION ANALYSIS
THE AFRICAN LION (Panthera leo leo): A CONTINENT-WIDE SPECIES DISTRIBUTION STUDY AND POPULATION ANALYSIS by Jason S. Riggio Dr. Stuart L. Pimm, Advisor May 2011 Masters project submitted in partial fulfillment of the requirements for the Master of Environmental Management degree in the Nicholas School of the Environment of Duke University 2011 Abstract Human population growth and land conversion across Africa makes the future of wide-ranging carnivores uncertain. For example, the African lion (Panthera leo leo) once ranged across the entire con- tinent – with the exception of the Sahara Desert and rainforests. It now lives in less than a quarter of its historic range. Recent research estimates a loss of nearly half of the lions in the past two decades. Some sources put their numbers as low as 20,000 individuals. Given these declines, conservation organ- izations propose to list the African lion as “endangered” under the U.S. Endangered Species Act and to upgrade the species’ CITES protections from Appendix II to Appendix I. To establish the lion’s current conservation status, I analyzed the size, distribution, and potential connections of populations across its range in Africa. It is particularly important to identify connected sub-populations and areas that can serve as corridors between existing protected areas. I compile the most current scientific literature, comparing sources to identify a current population estimate. I also use these sources to map known lion populations, potential habitat patches, and the connections between them. Finally, I assess the long-term viability of each lion population and determine which qualify as “lion strongholds.” The lion population assessment in this study has shown that over 30,000 lions remain in approx- imately 3,000,000 km2 of Africa. -

Lion (Panthera Leo Melanochaita) Diet in Relation to Prey Preference and Density in Meru National Park, Kenya
Lion (Panthera leo melanochaita) diet in relation to prey preference and density in Meru National Park, Kenya MSc Research Project Report 2019 – 2020 Mateo Bal MSc Biodiversity, Conservation and Restoration Evolutionary Ecology Group Department of Biology Institute of Environmental Sciences Antwerp University Leiden University Research supervisors: Prof. Dr. Ir. Hans H. de Iongh (Leiden University, Leo Foundation) MSc Luka L. Narisha (Kenya Wildlife Service) Lion (Panthera leo melanochaita) diet in relation to prey preference and density in Meru National Park, Kenya Mateo Bal s0185753 This research project was developed within a collaborative framework between the University of Antwerp, the Institute of Environmental Sciences of Leiden University, Kenya Wildlife Service, the Leo Foundation and the Born Free Foundation. Submitted to obtain the master’s degree in biology - specialisation Biodiversity, Conservation and Restoration. 2 ABSTRACT The African lion (Panthera leo) plays a key role in savannah ecosystems by directly and indirectly regulating trophic structure. Their foraging behavior has frequently been described as opportunistic, but often reveals a distinct preference for certain prey species that are energetically more profitable. This research project focussed on the population structure and diet of lions in Kenya’s Meru National Park. Data were collected from February until April 2019 and contribute to the PhD research of MSc Luka Narisha. A total of 28 lions were identified during fieldwork, indicating a lion density of 2.2 adult lions per 100 km2. Transect counts of potential prey species in the park revealed that Kirk’s dik-dik (Madoqua kirkii) had the highest relative abundance of all prey species (50.89%), while African buffalo (Syncerus caffer) contributed the most to the total prey biomass (33.94%). -

Threat of Rapid Extermination of the Lion (Panthera Leo Leo) in Waza National Park, Northern Cameroon
Threat of rapid extermination of the lion (Panthera leo leo) in Waza National Park, Northern Cameroon P. N. Tumenta1,2*, J. S. Kok1, J. C. van Rijssel1, R. Buij1,2, B. M. Croes1,2, P. J. Funston3, H. H. de Iongh1 and H. A. Udo de Haes1 1Institute of Environmental Sciences, Leiden University, PO Box 9518, 2300RA Leiden, The Netherlands, 2Centre for Environment and Development studies in Cameroon, Maroua ⁄ Department of Forestry, University of Dschang, PO Box 479, Dschang, Cameroon and 3Department of Nature Conservation, Tshwane University of Technology Pretoria, Private Bag X680 Pretoria, 0001, South Africa en identifiant individuellement ses membres. On a estime´ Abstract que cette population comptait de 14 a` 21 adultes. La Lion populations in West and Central Africa are small and structure d’aˆge e´tait biaise´e en faveur des adultes; les fragmented. In areas where park management is weak, lionceaux repre´sentaient 22% de tous les lions identifie´set threats will likely facilitate the extinction of the lion. le sex-ratio e´tait de 1 ⁄ 3. Deux des quatre lions e´quipe´s d’un Wildlife management requires knowledge of the popula- collier e´metteur furent tue´s ille´galement en un an, en guise tion estimate. The population of lions in Waza National de repre´sailles; deux autres maˆles et une autre femelle Park (Waza NP) was assessed by individual identification furent aussi tue´s pendant cette pe´riode. La population de of members in the population. The population was assessed lions semble avoir de´cline´ au cours des cinq dernie`res to comprise of 14–21 adult individual lions. -

African Lion (Panthera Leo) Behavior, Monitoring, and Survival in Human-Dominated Landscapes by Stephanie Dolrenry a Dissertatio
African lion (Panthera leo) behavior, monitoring, and survival in human-dominated landscapes By Stephanie Dolrenry A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Environment and Resources) at the UNIVERSITY OF WISCONSIN-MADISON 2013 Date of final oral examination: 8th January, 2013 The dissertation is approved by the following members of the Final Oral Committee: Dr. Robert S. Lutz, Associate Professor, Forest and Wildlife Ecology Dr. Calvin B. DeWitt, Professor Emeritus, Environmental Studies Dr. Laurence G. Frank, Associate Research Zoologist, Museum of Vertebrate Zoology Dr. Charles T. Snowdon, Professor, Psychology Dr. Stephen J. Ventura, Professor, Soil Science and Environmental Studies © Copyright by Stephanie Dolrenry 2013 All Rights Reserved i ACKNOWLEDGEMENTS “When spider webs unite, they can tie up a lion.” -Ethiopian Proverb This dissertation is the culmination of many people’s effort and support. First, thank you to the Kenyan Government and the Kenyan Wildlife Service (KWS), in particular the KWS unit of the Amboseli region, without their permissions this study would not have been possible. Secondly, thank you to the Maasai leaders and communities of Mbirikani, Eselenkei and Olgulului; it was only because of their blessings that this study was achieved. Thank you to R. Bonham and MPT/Big Life Foundation for your contribution and for your dedication to conservation on community lands. Thank you to Amboseli Trust for Elephants, Gamewatchers Porini, Amboseli Baboon Research Project, and Amboseli Lion Project; it is amazing to have committed neighbors who open their camps and share ideas. This work was made possible by the financial support of Panthera Foundation and the National Science Foundation; thank you for believing in this research. -

Amreli Index
DISTRICT PROFILE AMRELI INDEX 1 Amreli: A Snapshot 2 Economy and Industry Profile 3 Industrial Locations / Infrastructure 4 Support Infrastructure 5 Social Infrastructure 6 Tourism 7 Investment Opportunities 8 Annexure 2 1 Amreli: A Snapshot 3 Introduction: Amreli § Amreli is located near the Gulf of Khambhat in the Arabian Map1: District Map of Amreli with Talukas Sea, in the western part of Gujarat § The district has 11 talukas, of which the major ones are Amreli, Babra, Bagasara, Jafrabad, Rajula, Savarkundla and Vadiya § Amreli city is the district headquarter of Amreli § Focus industry sectors-Engineering, Port and Ship building, Minerals and Cement PipavavPort: Babra § Pipavav, India’s first private sector port and the world’s third Lathi largest container terminal operating port, is located in Amreli Vadia Amreli Lilia district (Source: Pipavav Port, 2007) Bagasara Savarkundla Minerals: Dhari Khambha Rajula § The district houses large reserves of limestone (Source: District Headquarter Jafrabad Mineral Treasure of Gujarat,, Commissioner of Geology and Mining, Talukas 2002-03) Pipavav City City 4 Fact File 70.30ºto 71.75ºEast (Longitude) Geographical location 20.45ºto 22.25ºNorth (Latitude) Average rainfall 550 mm Rivers Shetrunji, Thebi, Dhatarwadi, Gagdio, Shanti, Vadi and Rayadi Area 7,397 sq.km District headquarter Amreli Talukas 11 Population 1.39 million (As per 2001 Census) Population density 188 persons per sq. km. Sex ratio 987 Females per 1000 Males Literacy rate 66.10% Languages Gujarati, Hindi and English Seismic zone Zone III Source: Socio Economic Review 2006-07 5 2 Economy and Industry Profile 6 Economy and Industry Profile § Amreli is a base for cement, metallurgical, electrical equipments, ports and ship building industries § Due to the presence of large reserves of limestone, several major cement conglomerates, such as Larson & Toubro and Ultra Tech Cement Co.