Minimal Residual Disease Detection Using a Plasma-Only Circulating Tumor DNA Assay in Colorectal Cancer Patients
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Communicable Disease Chart
COMMON INFECTIOUS ILLNESSES From birth to age 18 Disease, illness or organism Incubation period How is it spread? When is a child most contagious? When can a child return to the Report to county How to prevent spreading infection (management of conditions)*** (How long after childcare center or school? health department* contact does illness develop?) To prevent the spread of organisms associated with common infections, practice frequent hand hygiene, cover mouth and nose when coughing and sneezing, and stay up to date with immunizations. Bronchiolitis, bronchitis, Variable Contact with droplets from nose, eyes or Variable, often from the day before No restriction unless child has fever, NO common cold, croup, mouth of infected person; some viruses can symptoms begin to 5 days after onset or is too uncomfortable, fatigued ear infection, pneumonia, live on surfaces (toys, tissues, doorknobs) or ill to participate in activities sinus infection and most for several hours (center unable to accommodate sore throats (respiratory diseases child’s increased need for comfort caused by many different viruses and rest) and occasionally bacteria) Cold sore 2 days to 2 weeks Direct contact with infected lesions or oral While lesions are present When active lesions are no longer NO Avoid kissing and sharing drinks or utensils. (Herpes simplex virus) secretions (drooling, kissing, thumb sucking) present in children who do not have control of oral secretions (drooling); no exclusions for other children Conjunctivitis Variable, usually 24 to Highly contagious; -

797 Circulating Tumor DNA and Circulating Tumor Cells for Cancer
Medical Policy Circulating Tumor DNA and Circulating Tumor Cells for Cancer Management (Liquid Biopsy) Table of Contents • Policy: Commercial • Coding Information • Information Pertaining to All Policies • Policy: Medicare • Description • References • Authorization Information • Policy History • Endnotes Policy Number: 797 BCBSA Reference Number: 2.04.141 Related Policies Biomarkers for the Diagnosis and Cancer Risk Assessment of Prostate Cancer, #336 Policy1 Commercial Members: Managed Care (HMO and POS), PPO, and Indemnity Plasma-based comprehensive somatic genomic profiling testing (CGP) using Guardant360® for patients with Stage IIIB/IV non-small cell lung cancer (NSCLC) is considered MEDICALLY NECESSARY when the following criteria have been met: Diagnosis: • When tissue-based CGP is infeasible (i.e., quantity not sufficient for tissue-based CGP or invasive biopsy is medically contraindicated), AND • When prior results for ALL of the following tests are not available: o EGFR single nucleotide variants (SNVs) and insertions and deletions (indels) o ALK and ROS1 rearrangements o PDL1 expression. Progression: • Patients progressing on or after chemotherapy or immunotherapy who have never been tested for EGFR SNVs and indels, and ALK and ROS1 rearrangements, and for whom tissue-based CGP is infeasible (i.e., quantity not sufficient for tissue-based CGP), OR • For patients progressing on EGFR tyrosine kinase inhibitors (TKIs). If no genetic alteration is detected by Guardant360®, or if circulating tumor DNA (ctDNA) is insufficient/not detected, tissue-based genotyping should be considered. Other plasma-based CGP tests are considered INVESTIGATIONAL. CGP and the use of circulating tumor DNA is considered INVESTIGATIONAL for all other indications. 1 The use of circulating tumor cells is considered INVESTIGATIONAL for all indications. -

Study Guide Medical Terminology by Thea Liza Batan About the Author
Study Guide Medical Terminology By Thea Liza Batan About the Author Thea Liza Batan earned a Master of Science in Nursing Administration in 2007 from Xavier University in Cincinnati, Ohio. She has worked as a staff nurse, nurse instructor, and level department head. She currently works as a simulation coordinator and a free- lance writer specializing in nursing and healthcare. All terms mentioned in this text that are known to be trademarks or service marks have been appropriately capitalized. Use of a term in this text shouldn’t be regarded as affecting the validity of any trademark or service mark. Copyright © 2017 by Penn Foster, Inc. All rights reserved. No part of the material protected by this copyright may be reproduced or utilized in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage and retrieval system, without permission in writing from the copyright owner. Requests for permission to make copies of any part of the work should be mailed to Copyright Permissions, Penn Foster, 925 Oak Street, Scranton, Pennsylvania 18515. Printed in the United States of America CONTENTS INSTRUCTIONS 1 READING ASSIGNMENTS 3 LESSON 1: THE FUNDAMENTALS OF MEDICAL TERMINOLOGY 5 LESSON 2: DIAGNOSIS, INTERVENTION, AND HUMAN BODY TERMS 28 LESSON 3: MUSCULOSKELETAL, CIRCULATORY, AND RESPIRATORY SYSTEM TERMS 44 LESSON 4: DIGESTIVE, URINARY, AND REPRODUCTIVE SYSTEM TERMS 69 LESSON 5: INTEGUMENTARY, NERVOUS, AND ENDOCRINE S YSTEM TERMS 96 SELF-CHECK ANSWERS 134 © PENN FOSTER, INC. 2017 MEDICAL TERMINOLOGY PAGE III Contents INSTRUCTIONS INTRODUCTION Welcome to your course on medical terminology. You’re taking this course because you’re most likely interested in pursuing a health and science career, which entails proficiencyincommunicatingwithhealthcareprofessionalssuchasphysicians,nurses, or dentists. -

Medical Microbiology and Infectious Diseases 22% Specialists in 2017 = 11%3
Medical Microbiology & Infectious Diseases Profile Updated December 2019 1 Table of Contents Slide . General Information 3-5 . Total number & number/100,000 population by province, 2019 6 . Number/100,000 population, 1995-2019 7 . Number by gender & year, 1995-2019 8 . Percentage by gender & age, 2019 9 . Number by gender & age, 2019 10 . Percentage by main work setting, 2019 11 . Percentage by practice organization, 2017 12 . Hours worked per week (excluding on-call), 2019 13 . On-call duty hours per month, 2019 14 . Percentage by remuneration method 15 . Professional & work-life balance satisfaction, 2019 16 . Number of retirees during the three year period of 2016-2018 17 . Employment situation, 2017 18 . Links to additional resources 19 2 General information Microbiology and infectious diseases focuses on the diagnosis and treatment of infectious diseases; thus, it is concerned with human illness due to micro-organisms. Since such disease can affect any and all organs and systems, this specialist must be prepared to deal with any region of the body. The specialty of Medical Microbiology and Infectious Disease consists primarily of four major spheres of activity: 1. the provision of clinical consultations on the investigation, diagnosis and treatment of patients suffering from infectious diseases; 2. the establishment and direction of infection control programs across the continuum of care; 3. public health and communicable disease prevention and epidemiology; 4. the scientific and administrative direction of a diagnostic microbiology laboratory. Source: Pathway evaluation program 3 General information Once you’ve completed medical school, it takes an additional 5 years of Royal College-approved residency training to become certified in medical microbiology and infectious disease. -

Diagnostic Tests Diagnostic Tests Attempt to Classify Whether Somebody Has a Disease Or Not Before Symptoms Are Present
community project encouraging academics to share statistics support resources All stcp resources are released under a Creative Commons licence stcp-rothwell-diagnostictests Diagnostic Tests Diagnostic tests attempt to classify whether somebody has a disease or not before symptoms are present. We are interested in detecting the disease early, while it is still curable. However, there is a need to establish how good a diagnostic test is in detecting disease. In this situation a 2x2 table similar to the below would be produced to test how effective a diagnostic test is at predicting the outcome of interest. A number of different measures can be calculated from this information. True Diagnosis Disease +ve Disease -ve Total Test Results +ve a b a+b -ve c d c+d a+c b+d N = This is the proportion of diseased individuals that are correctly Sensitivity: ( ) identified by the test as having the disease. (+ | ) + = This is the proportion of non-diseased individuals that Specificity: ( ) are correctly identified by the test as not having the disease. ( | ) + = This is the proportion − of individuals with Positive Predictive Value**: ( ) positive test results that are correctly diagnosed and actually have the disease. ( | + ) + = This is the proportion of individuals with Negative Predictive Value**: ( ) negative test results that are correctly diagnosed and do not have the disease. ( | + ) − © Joanne Rothwell Reviewer: Chris Knox University of Sheffield University of Sheffield Medical Statistics – diagnostic tests Positive Likelihood Ratio: = This gives a ratio of the test being positive + for patients with disease compared with those without disease. Aim to be much greater − than 1 for a good test. -

Burden of Disease: What Is It and Why Is It Important for Safer Food?
Burden of disease: what is it and why is it important for safer food? What is ‘burden of disease’? Burden of disease is concept that was developed in the 1990s by the Harvard School of Public Health, the World Bank and the World Health Organization (WHO) to describe death and loss of health due to diseases, injuries and risk factors for all regions of the world.1 The burden of a particular disease or condition is estimated by adding together: • the number of years of life a person loses as a consequence of dying early because of the disease (called YLL, or Years of Life Lost); and • the number of years of life a person lives with disability caused by the disease (called YLD, or Years of Life lived with Disability). Adding together the Years of Life Lost and Years of Life lived with Disability gives a single-figure estimate of disease burden, called the Disability Adjusted Life Year (or DALY). One DALY represents the loss of one year of life lived in full health (see text box for more explanation and examples). Looking at burden of disease using DALYs can reveal surprises about a population’s health. For example, the Global Burden of Disease 2004 Update suggests that neuropsychiatric conditions are the most important causes of disability in all regions of the world, accounting for around 33% of all years lived with disability among adults aged 15 years and over, but only 2.2% of deaths.2 Therefore while psychiatric disorders are not traditionally regarded as having a major impact on the health of populations, this picture is completely altered when the burden of disease is estimated using DALYs. -

Diabetic Foot and Gangrene
11 Diabetic Foot and Gangrene Jude Rodrigues and Nivedita Mitta Department of Surgery, Goa Medical College, India 1. Introduction “Early intervention in order to prevent potential disaster in the management of Diabetic foot is not only a great responsibility, but also a great opportunity” Despite advances in our understanding and treatment of diabetes mellitus, diabetic foot disease still remains a terrifying problem. Diabetes is recognized as the most common cause of non-traumatic lower limb amputation in the western world, with individuals over 20 times more likely to undergo an amputation compared to the rest of the population. There is growing evidence that the vascular contribution to diabetic foot disease is greater than was previously realised. This is important because, unlike peripheral neuropathy, Peripheral Arterial Occlusive Disease (PAOD) due to atherosclerosis, is generally far more amenable to therapeutic intervention. PAOD, has been demonstrated to be a greater risk factor than neuropathy in both foot ulceration and lower limb amputation in patients with diabetes. Diabetes is associated with macrovascular and microvascular disease. The term peripheral vascular disease may be more appropriate when referring to lower limb tissue perfusion in diabetes, as this encompasses the influence of both microvascular dysfunction and PAOD. Richards-George P. in his paper about vasculopathy on Jamaican diabetic clinic attendees showed that Doppler measurements of ankle/brachial pressure index (A/BI) revealed that 23% of the diabetics had peripheral occlusive arterial disease (POAD) which was mostly asymptomatic. This underscores the need for regular Doppler A/BI testing in order to improve the recognition, and treatment of POAD. -
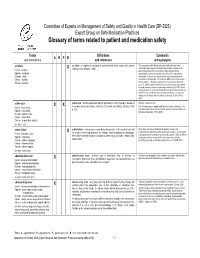
Glossary of Terms Related to Patient and Medication Safety
Committee of Experts on Management of Safety and Quality in Health Care (SP-SQS) Expert Group on Safe Medication Practices Glossary of terms related to patient and medication safety Terms Definitions Comments A R P B and translations and references and synonyms accident accident : an unplanned, unexpected, and undesired event, usually with adverse “For many years safety officials and public health authorities have Xconsequences (Senders, 1994). discouraged use of the word "accident" when it refers to injuries or the French : accident events that produce them. An accident is often understood to be Spanish : accidente unpredictable -a chance occurrence or an "act of God"- and therefore German : Unfall unavoidable. However, most injuries and their precipitating events are Italiano : incidente predictable and preventable. That is why the BMJ has decided to ban the Slovene : nesreča word accident. (…) Purging a common term from our lexicon will not be easy. "Accident" remains entrenched in lay and medical discourse and will no doubt continue to appear in manuscripts submitted to the BMJ. We are asking our editors to be vigilant in detecting and rejecting inappropriate use of the "A" word, and we trust that our readers will keep us on our toes by alerting us to instances when "accidents" slip through.” (Davis & Pless, 2001) active error X X active error : an error associated with the performance of the ‘front-line’ operator of Synonym : sharp-end error French : erreur active a complex system and whose effects are felt almost immediately. (Reason, 1990, This definition has been slightly modified by the Institute of Medicine : “an p.173) error that occurs at the level of the frontline operator and whose effects are Spanish : error activo felt almost immediately.” (Kohn, 2000) German : aktiver Fehler Italiano : errore attivo Slovene : neposredna napaka see also : error active failure active failures : actions or processes during the provision of direct patient care that Since failure is a term not defined in the glossary, its use is not X recommended. -

Molecular Pathological Epidemiology Gives Clues to Paradoxical Findings
Molecular Pathological Epidemiology Gives Clues to Paradoxical Findings The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Nishihara, Reiko, Tyler J. VanderWeele, Kenji Shibuya, Murray A. Mittleman, Molin Wang, Alison E. Field, Edward Giovannucci, Paul Lochhead, and Shuji Ogino. 2015. “Molecular Pathological Epidemiology Gives Clues to Paradoxical Findings.” European Journal of Epidemiology 30 (10): 1129–35. https://doi.org/10.1007/ s10654-015-0088-4. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:41392032 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Open Access Policy Articles, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#OAP HHS Public Access Author manuscript Author Manuscript Author ManuscriptEur J Epidemiol Author Manuscript. Author Author Manuscript manuscript; available in PMC 2016 October 07. Published in final edited form as: Eur J Epidemiol. 2015 October ; 30(10): 1129–1135. doi:10.1007/s10654-015-0088-4. Molecular Pathological Epidemiology Gives Clues to Paradoxical Findings Reiko Nishiharaa,b,c, Tyler J. VanderWeeled,e, Kenji Shibuyac, Murray A. Mittlemand,f, Molin Wangd,e,g, Alison E. Fieldd,g,h,i, Edward Giovannuccia,d,g, Paul Lochheadi,j, and Shuji Oginob,d,k aDepartment of Nutrition, Harvard T.H. Chan School of Public Health, 655 Huntington Ave., Boston, Massachusetts 02115 USA bDepartment of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, 450 Brookline Ave., Boston, Massachusetts 02215 USA cDepartment of Global Health Policy, Graduate School of Medicine, The University of Tokyo, 7-3-1, Hongo, Bunkyo-ku, Tokyo, Japan dDepartment of Epidemiology, Harvard T.H. -

Wisconsin Childhood Communicable Diseases Wall Chart
WISCONSIN CHILDHOOD COMMUNICABLE DISEASES Disease Name Incubation Period Time Period When Person is Spread by Signs and Symptoms Criteria for Exclusion from School or Group Onsite Control and Prevention Measures Time from exposure to Contagious (AKA, causative agent) symptoms Cold sores Direct contact with open sores Fever, irritability, blisters in mouth, on gums, lips, 2-7 weeks after symptoms appear, virus Exclude until fever-free, child able to control drooling, (Herpes simplex virus) or saliva 2 days to 2 weeks conjunctivitis, keratitis shedding possible without symptoms blisters resolved Mononucleosis Person to person contact with Many months after infection; excretion None, unless illness prevents participation; no contact saliva 30-50 days Fever, sore throat, swollen lymph nodes, fatigue of virus can occur intermittently for life sports until spleen no longer enlarged (Mono, Epstein-Barr virus) For all diseases: Good handwashing and hygiene; avoid Mumps R/V Inhalation of respiratory Fever, swelling and tenderness of parotid glands, Exclude for 5 days after swelling onset (day of swelling kissing, sharing drinks, or utensils, use proper disinfection droplets, direct contact with 12-25 days; headache, earache, painful swollen testicles, From 2 days before to 5 days after onset is day zero); exclude susceptible* contacts from of surfaces and toys (Mumps virus) saliva of infected person usually 16-18 days abdominal pain with swollen ovaries swelling day 12 through day 25 after exposure Mumps: Provide immunization records for exposed -

Integration of Molecular Pathology, Epidemiology and Social Science for Global Precision Medicine
Expert Review of Molecular Diagnostics ISSN: 1473-7159 (Print) 1744-8352 (Online) Journal homepage: http://www.tandfonline.com/loi/iero20 Integration of molecular pathology, epidemiology and social science for global precision medicine Akihiro Nishi, Danny A Milner Jr, Edward L Giovannucci, Reiko Nishihara, Andy S Tan, Ichiro Kawachi & Shuji Ogino To cite this article: Akihiro Nishi, Danny A Milner Jr, Edward L Giovannucci, Reiko Nishihara, Andy S Tan, Ichiro Kawachi & Shuji Ogino (2015): Integration of molecular pathology, epidemiology and social science for global precision medicine, Expert Review of Molecular Diagnostics, DOI: 10.1586/14737159.2016.1115346 To link to this article: http://dx.doi.org/10.1586/14737159.2016.1115346 Published online: 04 Dec 2015. Submit your article to this journal Article views: 82 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=iero20 Download by: [University of California, San Francisco] Date: 31 December 2015, At: 13:21 Perspectives Integration of molecular pathology, epidemiology and social science for global precision medicine Expert Rev. Mol. Diagn. Early online, 1–13 (2015) Akihiro Nishi1,2, The precision medicine concept and the unique disease principle imply that each patient has Danny A Milner Jr3,4, unique pathogenic processes resulting from heterogeneous cellular genetic and epigenetic Edward L alterations and interactions between cells (including immune cells) and exposures, including Giovannucci5,6,7, dietary, environmental, microbial and lifestyle factors. As a core method field in population health science and medicine, epidemiology is a growing scientific discipline that can analyze Reiko Nishihara5,6,8,9, 9,10 disease risk factors and develop statistical methodologies to maximize utilization of big data Andy S Tan , on populations and disease pathology. -

Autopsy Protocol
140 Part 4 Records and Reports Section 404.3 Case Files Regardless of whether you develop investigative protocols it is incumbent upon you to maintain as thorough and organized set of investigative files as possible. The investigative files should include but not be restricted to the following: all reports, investigator's notes, sketches and death scene photographs, reports of autopsy and laboratory analyses of evidence, copies of all forms completed by the coroner to include chain-of-custody forms and laboratory request forms. The major objective is to maintain as complete and proper file as possible. Section 501 Autopsy Protocol Section 501.1 Overview.............................................................................................................................. 115 Section 501.2 Ordering an Autopsy............................................................................................................ 115 Section 501.3 Autopsy Protocol.................................................................................................................. 117 Section 501.1 Overview Coroners may find the following definition of forensic pathology useful to their work. Forensic pathology is the branch of medical practice that produces evidence useful in the criminal justice administration, public health and public safety. Under this definition are three key elements: Cause of Death, Manner of Death and Mechanism of Death. The cause of death related to the disease, injury or abnormality that alone or together in some combination