Mouse Genome Monthly
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional HCL Notes Email Archiving Solution
AM ™ ArchiveMail DATASHEET Functional HCL Notes™ email archiving solution Key In Detail on Introducti Characteristics Benefits For the Users: Continuous growth in data volumes related to messaging and the practical limits imposed by Notes database architecture make email archiving systems essential. M Improved productivity due to automatic management of archive. COOPERTEAM’s Archive Mail™ is a functional archiving solution which responds to the M needs of administrators by allowing them to manage archiving policies efficiently and Access to archived documents from automatically, whilst still guaranteeing their users access to their documents and the Notes client, iNotes and mobile attached files. devices. Archive Mail™ automates the archiving of messages, calendar entries, tasks and contacts M Multi-criteria searching available for to one archive per year, hosted on either dedicated or shared function servers. Archiving both server & workstation. policies can be based on document ageing criteria. In addition, the archive process can be triggered when the mail database exceeds its For the Administrators: threshold (quota), or manually by users after selecting documents to be archived. M Optimization of the Domino Archive Mail™ allows administrators to manage multiple archive databases and easily infrastructure. implement ILM (Information Lifecycle Management) policies, and to correctly align the M Align messaging storage costs, value storage costs of the archived documents with their value to the business. for the business and quota policies. M Once Archive Mail™ is implemented, documents that meet the configured ageing Automated and centralised parameters for archiving are transferred to an archive database at a frequency predeter- management of archiving mined by their profile administrator. -

Zimbra Tips & Tricks
TOP ZIMBRA 15 TIPS & TRICKS This guide brings you the top 15 Zimbra Tips & Tricks to help you save time while working in Zimbra. If you find these Tips & Tricks helpful, you may also like our tutorial demos: The top 15 Zimbra Tips & Tricks (based on clicks on Zimbra’s “Did You Know?” blog) are: Throughout this guide, look for pop-up boxes highlighting quick tips Email Tips and Tricks and facts. 1 Searching in Zimbra TIP: Skip ahead to any 2 Importing Into and Exporting From Zimbra section of the 15 Tips & 3 Changing Your Zimbra Preferences Tricks by clicking on the 4 Changing Your Zimbra Theme titles to the right. 5 Setting Zimbra Email and SMS (text) Reminders 6 Enabling Zimbra’s Mandatory Spellcheck 7 Sorting Email 8 Creating an Appointment with the Mini Calendar 9 Creating an Email Distribution List or Contact Group 10 Opening an Email in a Separate Window 11 Turning on Zimbra’s Activity Stream 12 Switching from Conversation View to Message View 13 Sharing Your Zimbra Calendar Outside Your Organization 14 Sharing in Zimbra 15 Enabling Zimbra Two-Factor Authentication 1 Searching in Zimbra There are many great and easy ways to search in Zimbra: • Quick keyword search • Complex search • Right-click search Quick Keyword Search You can do a quick search using keywords like from, to, in, etc. Click here to see all search keywords. How much time do you spend searching To search by keyword: for a specific email? keyword: search word(s) Searches are easy and fast in Zimbra. Search Examples: your entire Zimbra • Find all email received from Mark (all email with “Mark” in the From account, including attachments, in seconds. -

The History of Spam Timeline of Events and Notable Occurrences in the Advance of Spam
The History of Spam Timeline of events and notable occurrences in the advance of spam July 2014 The History of Spam The growth of unsolicited e-mail imposes increasing costs on networks and causes considerable aggravation on the part of e-mail recipients. The history of spam is one that is closely tied to the history and evolution of the Internet itself. 1971 RFC 733: Mail Specifications 1978 First email spam was sent out to users of ARPANET – it was an ad for a presentation by Digital Equipment Corporation (DEC) 1984 Domain Name System (DNS) introduced 1986 Eric Thomas develops first commercial mailing list program called LISTSERV 1988 First know email Chain letter sent 1988 “Spamming” starts as prank by participants in multi-user dungeon games by MUDers (Multi User Dungeon) to fill rivals accounts with unwanted electronic junk mail. 1990 ARPANET terminates 1993 First use of the term spam was for a post from USENET by Richard Depew to news.admin.policy, which was the result of a bug in a software program that caused 200 messages to go out to the news group. The term “spam” itself was thought to have come from the spam skit by Monty Python's Flying Circus. In the sketch, a restaurant serves all its food with lots of spam, and the waitress repeats the word several times in describing how much spam is in the items. When she does this, a group of Vikings in the corner start a song: "Spam, spam, spam, spam, spam, spam, spam, spam, lovely spam! Wonderful spam!" Until told to shut up. -

Guest Artist Agreement
Artists Agreement Unitarian Universalist Congregation of Phoenix 4027 E. Lincoln Drive, Paradise Valley, AZ 85253 Phone: 602-840-8400 Email: [email protected] The Sanctuary Art Team of the Unitarian Universalist Congregation of Phoenix (UUCP) welcomes you. We are pleased that you are participating in our art exhibit. The following guidelines and liability agreement should be helpful to you in preparing for installation of your artwork. 1) All works selected for exhibit will be displayed in the Sanctuary or Hall in spaces reserved for that purpose. 2) An art inventory should be completed before arrival and sent to the email above or as directed. It MUST be completed when the art is delivered to UUCP. Artists must provide the following information: Artist's Name, Title of Work, Medium, Dimensions, and Sale Price or NFS (See inventory form). 3) All work must be ready for hanging. Any work over 600 square inches requiring protective covering must be covered in Plexiglas (NOT glass). 4) All works must remain on display until the termination of the show unless previously arranged with the Sanctuary Art Team. At the close of the show, the lender/artist agrees to remove the pieces on exhibit or arrange for their removal. Any object remaining in the Sanctuary for 30 days following the scheduled close of the show will be handled at the discretion of the Sanctuary Art Team. 5) Each object displayed will be exhibited in such manner as the Sanctuary Art Team decides. 6) The Sanctuary Art Team reserves the right to select and/or reject works, bearing in mind that the Sanctuary is not a conventional gallery and the selected works must be appropriate for all congregational activities. -

Monthly Worship Themes What Does It Mean to Be a People Of
2 Begin Your Journey Here Sunday Worship Monthly Worship Themes What does it mean to be a people of... Welcome to UUCP! This is a religious community September Covenant Welcoming all as we Rev. Susan October Freedom Share our Journeys Frederick-Gray Grow in Spirit November Gratitude Together work to Advance Justice December Tradition The best way to see, feel and get to know this community January Truth is through Sunday morning services. Each month we February Love explore a theme in depth. Our themes challenge us to grow in meaning, build connections across diversities and March Hope make our world a better place. April Service May Peace Service times: May – September 13: 10:30 AM June Strength September 20 – April 17: 9:30 AM and 11:15 AM July/August Spirituality Starting Point Community Nights Are you new to UUCP? Do you have questions Break Bread and Make Friends at UUCP. Community about UUCP and Unitarian Universalism? Nights are a time of fellowship and community building Starting Point is an informal program for all ages. We provide the main dish with a vegetarian especially for you! Meet a representative from option and one side. Just bring your appetite, enthusi- the Membership Committee by the Starting asm, and a side dish or dessert to share if able. You are Point sign in the front lobby any second Sunday of the also welcome to bring a sack supper if you prefer. month. You don’t have to sign up. Just show up! Chalice Kids is a program for children that begins right In twenty to thirty minutes, we will answer questions and after the meal at 6:30 PM. -
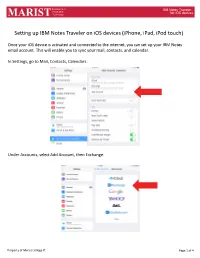
Setting up IBM Notes Traveler on Ios Devices (Iphone, Ipad, Ipod Touch)
Department of IBM Notes Traveler Information for iOS devices MARIST Technology Setting up IBM Notes Traveler on iOS devices (iPhone, iPad, iPod touch) Once your iOS device is activated and connected to the internet, you can set up your IBM Notes email account. This will enable you to sync your mail, contacts, and calendar. In Settings, go to Mail, Contacts, Calendars. Under Accounts, select Add Account, then Exchange. Property of Marist College IT Page 1 of 4 Department of IBM Notes Traveler Information for iOS devices MARIST Technology The Exchange window will display. In the Email field, enter your Marist email address. In the Password field, enter your IBM iNotes password (as you would log in to notes.marist.edu). In the Description field, type “Marist” (without quotation marks). Press “Next” in the upper right corner of the window. The window will expand. In the Server field type in “traveler.notes.marist.edu/traveler” (without quotation marks). In the Username field enter your iNotes username (firstname.lastname). Leave the Domain field blank. Press “Next” in the upper right corner of the window. Information Technology Property of Marist College IT Page 2 of 4 Department of IBM Notes Traveler MARIST Your account will be verified. The next screen displays the Notes data items that will Sync. Slide to the left any item you do not wish to sync, select Save. Information Technology for iOS devices Information Technology for iOS devices The Exchange Account will be created and you will be returned to Settings. Change the ‘Mail Days to Sync’ to your preference. -

How to Create a Personal Email Account/Address (Gmail, Yahoo & Outlook)
How to create a personal email account/address (Gmail, Yahoo & Outlook) Step 1 – select an email provider (Gmail, Yahoo, Outlook, etc.) There are many different email providers that will allow you to create an email account to send and receive emails. Some of the more common ones include Gmail, Yahoo and Outlook. Below are instructions for setting up an email account using these three providers. Gmail – 1) Go to www.gmail.com 2) Click create an account 3) You will get a small pop up box that asks you to select either: for myself or to manage my business. Select For myself 4) A form to create your account will appear. Complete the information in the form. a. First name, Last name b. Username – enter the first part of what you want your email address to be – for example, janedoe (@gmail.com will be added to the end of it.) For example: [email protected] c. Password – you will need to create a password. This is what will allow you to log into your account to send and receive emails 5) Next, you will see a form to enter some of your personal information, such as your phone number, birth date and gender – enter the requested information 6) You will receive Google’s Privacy and Terms. Read and click “I Agree” (you will need to agree to the privacy and terms in order to complete your email account sign up) 7) Once you agree to the terms and conditions, you will be taken to your new email account inbox 8) You can now start to send and receive emails 9) You can login to your new email account by going to www.gmail.com (and entering your email address and password) Yahoo – 1) Go to https://login.yahoo.com/account/create 2) Complete the information in the sign-up form that appears. -
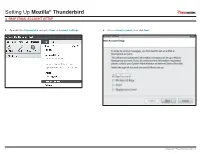
Setting up Mozilla® Thunderbird IMAP EMAIL ACCOUNT SETUP
Setting Up Mozilla® Thunderbird IMAP EMAIL ACCOUNT SETUP 1. Open Mozilla Thunderbird and go to Tools -> Account Settings. 2. Choose Email account; then click Next. 1 Copyright © EmailHosting.com LLC Setting Up Mozilla® Thunderbird IMAP EMAIL ACCOUNT SETUP (cont.) 3. In the Identity window type your name as you want it to appear on outgoing emails. 4. In the Server Information window select the radio button labeled IMAP. • Then, type your email address in the field labeled Email Address. • Enter imap.emailhosting.com in the field labeled Incoming Server. • Click Next to continue. • Enter smtp.emailhosting.com in the field labeled Outgoing Server. • Click Next to continue. John Smith imap.emailhosting.com [email protected] smtp.emailhosting.com Note: If you have already used Thunderbird in the past the Outgoing Server will not be listed. 2 Copyright © EmailHosting.com LLC Setting Up Mozilla® Thunderbird IMAP EMAIL ACCOUNT SETUP (cont.) 5. In the User Names window, the fields should already be filled out correctly. 6. Click Next on the Account Name page to accept the default Account Name. • Confirm that both fields match your EmailHosting username. • In the Congratulations window, verify that all information is correct. • Click Next to continue. • Then click Finish. [email protected] [email protected] [email protected] [email protected] 7. You will be prompted for your password. If you’d like to save your password, make sure Use Password Manager to remember this password is checked before clicking on OK to continue. Note: The field Outgoing Server is only visible during the setup of Mozilla Thunderbird’s first email account. -

Webaccess Client User Guide
GroupWise Web User Guide July 2021 Legal Notices © Copyright 1993 - 2021 Micro Focus or one of its affiliates. The only warranties for products and services of Micro Focus and its affiliates and licensors (“Micro Focus”) are set forth in the express warranty statements accompanying such products and services. Nothing herein should be construed as constituting an additional warranty. Micro Focus shall not be liable for technical or editorial errors or omissions contained herein. The information contained herein is subject to change without notice. Contents About This Book 5 1 Basic Tasks 7 Logging In . 7 Sending Items . 7 Reading Items. 8 Replying to Items . 8 Changing the Compose View . 8 Managing Signatures . 8 Using the Preview Pane . 9 Scheduling an Appointment . 9 Accepting Appointments . 9 Sending and Receiving Attachments . 10 Managing Folders and Items . 10 Creating a Folder . 10 Moving a Folder . 10 Searching for Items. 10 Managing Your Calendars. 11 Viewing Your Calendars . 11 Creating a New Calendar. 11 Viewing Your Contacts . 11 Managing Your Out of Office Rule. 11 2 Advanced Tasks 13 Using Rules. 13 Sharing Folders and Calendars . 13 Using Categories . 13 Viewing Message Details . 14 Marking Items Read/Unread/Private/Complete/Incomplete . .14 Using Proxy. 14 Contents 3 4 About This Book The GroupWise Web User Guide provides conceptual information about GroupWise Web. This book includes step-by-step guidance for common tasks. Intended Audience This book provides information for users wanting to use GroupWise in a web browser. Additional Documentation For the most recent version of this guide and other GroupWise documentation resources, visit the GroupWise 18 Documentation website (https://www.novell.com/documentation/groupwise18/). -

How Do I View My Email History? How Do I View My Email History?
Help Articles > Club Help > Club - Members' Guide > My ClubRunner > How do I view my email history? How do I view my email history? Omar S. - 2021-03-04 - My ClubRunner Tracking and viewing emails sent to you through ClubRunner is now easier than ever. With the My Email History page under the My ClubRunner tab in your Member Area you can see a list of all ClubRunner emails sent to your inbox, view the delivery statistics, and even open the email itself to see its contents. 1. To access your email history in ClubRunner, login to your Member Area and click on the My ClubRunner tab. 2. Next, select the My Email History tab below. 3. On this page, you can view email delivery stats as well as the number of emails that you have opened, and any undelivered or otherwise erroneous emails. 4. By Clicking on the View icon under Actions for any of the emails, you can see the content of the email directly on your Web Browser. 5. There are several different Statuses that an email address could have: Sent - The email was sent to the recipient. The email could have been opened, but not reported as opened due to anti virus blocking the notice of it being opened. Opened - The email was opened. This could be false, the anti virus could have opened it to check the email and the recipient never opened it. Or the anti virus or email client could have blocked the message coming back to us saying the email was opened. To learn more about email tracking please read the article about email tracking. -

Unitarian Universalist Congregation of Phoenix Justice Ministry Team Application Form
Unitarian Universalist Congregation of Phoenix Justice Ministry Team Application Form Team Responsibilities Each team shall have a designated leader to act as the communication point person. This person will receive important announcements from the office regarding deadlines and other congregation business matters. Additionally, this person will receive forwarded email and phone inquiries regarding this group. Teams shall maintain a record of participation for all events to include in their Annual Report. Submissions are due April 30. Teams shall submit a description of the group and a schedule of all activities for the upcoming year (August through July) to be included in the Program Catalog. Submissions are due May 31. Teams shall send a leader or delegate to the quarterly Council Meetings. Teams shall adhere to the publicity guidelines in the annual Guidebook for Volunteer and Paid Staff. Teams expire at the end of June on the second year after approval. If a team wants to continue, another application must be submitted by the end of March. If a team wants to request funding support from UUCP, they need to submit their budget request before the end of January so that it can go into the annual budget process. Application Process Application to start a new team may be made at any time. Please submit the application to the UUCP office, attention Rev. Susan, or email [email protected]. Please note that application approval may take up to six weeks. Unitarian Universalist Congregation of Phoenix Justice Ministry Team Application Form Team Leader or Team-Co-Leaders Must be UUCP Members Name: Phone #: Email: Name: Phone #: Email: Team Members List the names of at least four additional committed team members. -

Zimbra Web Client User Guide Advanced Web Client
Zimbra Web Client User Guide Advanced Web Client Zimbra Collaboration 8.7 Legal Notices Copyright © 2016 Synacor, Inc. All rights reserved. This product is protected by U.S. and international copyright and intellectual property laws. ZIMBRA is a trademark of Synacor, Inc. in the United States and/or other jurisdictions. All other marks and names mentioned herein may be trademarks of their respective companies. Synacor, Inc. 40 La Riviere Drive, Suite 300 Buffalo, New York 14202 www.synacor.com Zimbra Collaboration 8.7 February 2016 Table of Contents 1 Setting Up Your Account . .13 Using Advanced and Standard Versions . 13 Using the Touch Client . 13 Signing In and Signing Out . 14 Sign In . 14 Sign Out . 14 Choose a Default Client Version . 14 If Your Session Expires . 15 Changing Your Password . 15 Navigating the Zimbra Web Client . 16 Use Keyboard Shortcuts . 17 Use Browser Tools . 18 Get Help . 18 Personalizing Your Account . 18 Change the Theme . 18 Change Font and Size . 18 Change Your Default Time Zone . 18 Change the Language . 19 Change Text Direction . 19 Move the Reading Pane . 19 Change the Pane Size . 19 Accessing External Accounts . 19 Add an External Account . 20 Set Up POP Behavior for Your Account . 21 Using Exchange Web Services (EWS) . 22 Clients Supported for EWS . 22 Configure Mac Outlook . 22 Configure Mac Native Mail Client . 22 2 Managing Your Email . 23 Customizing Your Mailbox Initial View . 23 View Email Messages by Conversation . 24 Change the Message View . 24 Using Zimlets . 24 Enable or Disable a Zimlet . 24 Using A Reading Pane . 24 Turn Reading Pane On or Off .