Latitudinal Cline in Chromosome Numbers of Ice Cod A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Boreogadus Saida) and Safron Cod (Eleginus Gracilis) Early Life Stages in the Pacifc Arctic
Polar Biology https://doi.org/10.1007/s00300-019-02494-4 ORIGINAL PAPER Spatio‑temporal distribution of polar cod (Boreogadus saida) and safron cod (Eleginus gracilis) early life stages in the Pacifc Arctic Cathleen D. Vestfals1 · Franz J. Mueter2 · Janet T. Dufy‑Anderson3 · Morgan S. Busby3 · Alex De Robertis3 Received: 24 September 2018 / Revised: 15 March 2019 / Accepted: 18 March 2019 © Springer-Verlag GmbH Germany, part of Springer Nature 2019 Abstract Polar cod (Boreogadus saida) and safron cod (Eleginus gracilis) are key fshes in the Arctic marine ecosystem, serving as important trophic links between plankton and apex predators, yet our understanding of their life histories in Alaska’s Arctic is extremely limited. To improve our knowledge about their early life stages (ELS), we described the spatial and temporal distributions of prefexion larvae to late juveniles (to 65 mm in length) in the Chukchi and western Beaufort seas based on surveys conducted between 2004 and 2013, and examined how their abundances varied in response to environmental factors. Species-specifc diferences in habitat use were found, with polar cod having a more ofshore and northern distribution than safron cod, which were found closer inshore and farther south. Polar cod prefexion and fexion larvae were encountered throughout the sampling season across much of the shelf, which suggests that spawning occurs over several months and at multiple locations, with Barrow Canyon potentially serving as an important spawning and/or retention area. Polar cod ELS were abundant at intermediate temperatures (5.0–6.0 °C), while safron cod were most abundant at the highest temperatures, which suggests that safron cod may beneft from a warming Arctic, while polar cod may be adversely afected. -

The Minke Whale
Status of Marine Mammals in the North Atlantic THE MINKE WHALE This series of reports is intended to provide information on North Atlantic marine mammals suitable for the general reader. Reports are produced on species that have been considered by the NAMMCO Scientific Committee, and therefore reflect the views of the Council and Scientific Committee of NAMMCO. North Atlantic Marine Mammal Commission Polar Environmental Centre N-9296 Tromsø, Norway Tel.: +47 77 75 01 80, Fax: +47 77 75 01 81 Email: [email protected], Web site: www.nammco.no MINKE WHALE (Balaenoptera acutorostrata) The minke whale is the smallest of the balaenopterids, or rorquals. It attains a length of 8-9 m and a weight of about 8 tonnes in the North Atlantic. As with all balaenopterids, the females are somewhat larger than the males. Minke whales are black or dark grey dorsally and white on the ventral side. A transverse white band is charachteristic for the species in the Northern Hemisphere. With a worldwide distribution, it is the most common of the rorquals. Distribution and Stock Definition: The minke whale is found throughout most of the North Atlantic, but is generally more common in coastal or shelf areas (Fig. 1). Although the migratory patterns of North Atlantic minke whales are not known, they tend to occupy higher latitudes in the summer and lower latitudes in the winter. Breeding and calving areas are not known. North Atlantic minke whales have been divided into four management stocks by the International Whaling Commission (IWC) (Donovan 1991) (See Fig. 1). The original stock divisions were not based on extensive biological information. -

Complete Mitochondrial Genome Sequences of the Arctic Ocean Codwshes Arctogadus Glacialis and Boreogadus Saida Reveal Oril and Trna Gene Duplications
Polar Biol (2008) 31:1245–1252 DOI 10.1007/s00300-008-0463-7 ORIGINAL PAPER Complete mitochondrial genome sequences of the Arctic Ocean codWshes Arctogadus glacialis and Boreogadus saida reveal oriL and tRNA gene duplications Ragna Breines · Anita Ursvik · Marianne Nymark · Steinar D. Johansen · Dag H. Coucheron Received: 4 December 2007 / Revised: 16 April 2008 / Accepted: 5 May 2008 / Published online: 27 May 2008 © The Author(s) 2008 Abstract We have determined the complete mitochon- Introduction drial genome sequences of the codWshes Arctogadus gla- cialis and Boreogadus saida (Order Gadiformes, Family More than 375 complete sequenced mitochondrial genomes Gadidae). The 16,644 bp and 16,745 bp mtDNAs, respec- from ray-Wnned Wshes have so far (December 2007) been tively, contain the same set of 37 structural genes found in submitted to the database (http://www.ncbi.nlm.nih.gov), all vertebrates analyzed so far. The gene organization is and many of these sequences have contributed considerably conserved compared to other Gadidae species, but with one to resolving phylogenetic relationships among Wshes. Evo- notable exception. B. saida contains heteroplasmic rear- lutionary relationships at diVerent taxonomic levels have rangement-mediated duplications that include the origin of been addressed, including Division (Inoue et al. 2003; Miya light-strand replication and nearby tRNA genes. Examina- et al. 2003), Subdivision (Ishiguro et al. 2003), Genus tion of the mitochondrial control region of A. glacialis, (Doiron et al. 2002; Minegishi et al. 2005), and Species B. saida, and four additional representative Gadidae genera (Yanagimoto et al. 2004; Ursvik et al. 2007). identiWed a highly variable domain containing tandem The circular mitochondrial genomes from ray-Wnned repeat motifs in A. -

Balaenoptera Acutorostrata ) in Icelandic Waters 2003-2007
Paper 9 Víkingsson GA., Auðunsson GA., Elvarsson BT. and Gunnlaugsson T. (2013). Energy storage in common minke whales (Balaenoptera acutorostrata ) in Icelandic waters 2003-2007. -Chemical composition of tissues and organs. IWC. SC/F13/SP10. SC/F13/SP10 Energy storage in common minke whales (Balaenoptera acutorostrata ) in Icelandic waters 2003-2007 . – Chemical composition of tissues and organs. Gísli A. Víkingsson 1, Guðjón Atli Auðunsson 2, Bjarki Þór Elvarsson 1,3 , Þorvaldur Gunnlaugsson 1. 1Marine Research Institute, Skúlagata 4, IS-101 Reykjavík, Iceland 2Innovation Center Iceland, Dept.Ana.Chem., Árleynir 2-8, IS-112 Reykjavik, Iceland 3Science Institute, University of Iceland, Tæknigarður, Dunhagi 5, 107 Reykjavík Iceland Abstract This report details studies on chemical composition (total lipids, protein and water) of various tissues in common minke whales ( Balaenoptera acutorostrata ). Energy deposition was demonstrated by an increase in the percentage of lipids in blubber, muscle, visceral fat and bones. As in other balaenopterids, most lipids were deposited dorsally behind the dorsal fin. In addition, large amounts of energy are apparently stored as visceral fats and within bone tissue. Highest levels of lipids were found in pregnant females. Spatial variation within the Icelandic continental shelf area might be explained by corresponding variation in diet composition. A significant decrease over the research period 2003-2007 in lipid content of posterior dorsal muscle could be a result of a decrease in prey availability Introduction The common minke whale ( Balaenoptera acutorostrata ) is the most abundant baleen whale species in the Icelandic continental shelf area. Like other Balaenopterids, minke whales are migratory animals spending the summer at relatively high latitude feeding areas and the winters at lower latitude breeding areas (Horwood 1990). -

Seasonal Ecology in Ice-Covered Arctic Seas - Considerations for Spill Response Decision Making
Accepted Manuscript Seasonal ecology in ice-covered Arctic seas - Considerations for spill response decision making Magnus Aune, Ana Sofia Aniceto, Martin Biuw, Malin Daase, Stig Falk-Petersen, Eva Leu, Camilla A.M. Ottesen, Kjetil Sagerup, Lionel Camus PII: S0141-1136(17)30699-2 DOI: 10.1016/j.marenvres.2018.09.004 Reference: MERE 4594 To appear in: Marine Environmental Research Received Date: 14 November 2017 Revised Date: 9 March 2018 Accepted Date: 3 September 2018 Please cite this article as: Aune, M., Aniceto, A.S., Biuw, M., Daase, M., Falk-Petersen, S., Leu, E., Ottesen, C.A.M., Sagerup, K., Camus, L., Seasonal ecology in ice-covered Arctic seas - Considerations for spill response decision making, Marine Environmental Research (2018), doi: 10.1016/j.marenvres.2018.09.004. This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT 1 Seasonal ecology in ice-covered Arctic seas - considerations for spill 2 response decision making 3 Magnus Aune 1,6, Ana Sofia Aniceto 1,2 , Martin Biuw 3, Malin Daase 4, Stig Falk-Petersen 1,4, 4 Eva Leu 5, Camilla A.M. Ottesen 4, Kjetil Sagerup 1, Lionel Camus 1 5 6 1Akvaplan-niva -

Determination of Trophic Relationships Within a High Arctic Marine Food Web Using 613C and 615~ Analysis *
MARINE ECOLOGY PROGRESS SERIES Published July 23 Mar. Ecol. Prog. Ser. Determination of trophic relationships within a high Arctic marine food web using 613c and 615~ analysis * Keith A. ~obson'.2, Harold E. welch2 ' Department of Biology. University of Saskatchewan, Saskatoon, Saskatchewan. Canada S7N OWO Department of Fisheries and Oceans, Freshwater Institute, 501 University Crescent, Winnipeg, Manitoba, Canada R3T 2N6 ABSTRACT: We measured stable-carbon (13C/12~)and/or nitrogen (l5N/l4N)isotope ratios in 322 tissue samples (minus lipids) representing 43 species from primary producers through polar bears Ursus maritimus in the Barrow Strait-Lancaster Sound marine food web during July-August, 1988 to 1990. 613C ranged from -21.6 f 0.3%0for particulate organic matter (POM) to -15.0 f 0.7%0for the predatory amphipod Stegocephalus inflatus. 615~was least enriched for POM (5.4 +. O.8%0), most enriched for polar bears (21.1 f 0.6%0), and showed a step-wise enrichment with trophic level of +3.8%0.We used this enrichment value to construct a simple isotopic food-web model to establish trophic relationships within thls marine ecosystem. This model confirms a food web consisting primanly of 5 trophic levels. b13C showed no discernible pattern of enrichment after the first 2 trophic levels, an effect that could not be attributed to differential lipid concentrations in food-web components. Although Arctic cod Boreogadus saida is an important link between primary producers and higher trophic-level vertebrates during late summer, our isotopic model generally predicts closer links between lower trophic-level invertebrates and several species of seabirds and marine mammals than previously established. -

And Narwhals (Monodon Monoceros)In the Canadian High Arctic Determined by Stomach Content and Stable Isotope Analysis Jordan K
RESEARCH/REVIEW ARTICLE Foraging ecology of ringed seals (Pusa hispida), beluga whales (Delphinapterus leucas) and narwhals (Monodon monoceros)in the Canadian High Arctic determined by stomach content and stable isotope analysis Jordan K. Matley,1 Aaron T. Fisk2 & Terry A. Dick3 1 Centre for Sustainable Tropical Fisheries and Aquaculture, College of Marine and Environmental Sciences, James Cook University, Townsville, QLD 4811, Australia 2 Great Lakes Institute for Environmental Research, University of Windsor, Windsor, ON N9B 3P4, Canada 3 Department of Biological Sciences, University of Manitoba, Winnipeg, MB R3T 2N2, Canada Keywords Abstract Arctic marine mammals; stable isotopes; Stomach content and stable isotope analysis (d13C and d15N from liver and stomach contents; Bayesian mixing models; diet; Arctic cod. muscle) were used to identify habitat and seasonal prey selection by ringed seals (Pusa hispida; n21), beluga whales (Delphinapterus leucas; n13) Correspondence and narwhals (Monodon monoceros; n3) in the eastern Canadian Arctic. Jordan K. Matley, College of Marine Arctic cod (Boreogadus saida) was the main prey item of all three species. Diet and Environmental Sciences, James Cook reconstruction from otoliths and stable isotope analysis revealed that while University, Townsville, QLD 4811, Australia. ringed seal size influenced prey selection patterns, it was variable. Prey-size E-mail: [email protected] selection and on-site observations found that ringed seals foraged on smaller, non-schooling cod whereas belugas and narwhals consumed larger individuals in schools. Further interspecific differences were demonstrated by d13C and d15N values and indicated that ringed seals consumed inshore Arctic cod compared to belugas and narwhals, which foraged to a greater extent offshore. -

Biodiversity of Arctic Marine Fishes: Taxonomy and Zoogeography
Mar Biodiv DOI 10.1007/s12526-010-0070-z ARCTIC OCEAN DIVERSITY SYNTHESIS Biodiversity of arctic marine fishes: taxonomy and zoogeography Catherine W. Mecklenburg & Peter Rask Møller & Dirk Steinke Received: 3 June 2010 /Revised: 23 September 2010 /Accepted: 1 November 2010 # Senckenberg, Gesellschaft für Naturforschung and Springer 2010 Abstract Taxonomic and distributional information on each Six families in Cottoidei with 72 species and five in fish species found in arctic marine waters is reviewed, and a Zoarcoidei with 55 species account for more than half list of families and species with commentary on distributional (52.5%) the species. This study produced CO1 sequences for records is presented. The list incorporates results from 106 of the 242 species. Sequence variability in the barcode examination of museum collections of arctic marine fishes region permits discrimination of all species. The average dating back to the 1830s. It also incorporates results from sequence variation within species was 0.3% (range 0–3.5%), DNA barcoding, used to complement morphological charac- while the average genetic distance between congeners was ters in evaluating problematic taxa and to assist in identifica- 4.7% (range 3.7–13.3%). The CO1 sequences support tion of specimens collected in recent expeditions. Barcoding taxonomic separation of some species, such as Osmerus results are depicted in a neighbor-joining tree of 880 CO1 dentex and O. mordax and Liparis bathyarcticus and L. (cytochrome c oxidase 1 gene) sequences distributed among gibbus; and synonymy of others, like Myoxocephalus 165 species from the arctic region and adjacent waters, and verrucosus in M. scorpius and Gymnelus knipowitschi in discussed in the family reviews. -
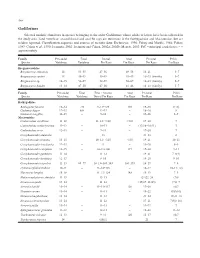
Gadiformes Selected Meristic Characters in Species Belonging to the Order Gadiformes Whose Adults Or Larvae Have Been Collected in the Study Area
548 Gadiformes Selected meristic characters in species belonging to the order Gadiformes whose adults or larvae have been collected in the study area. Total vertebrae, second dorsal and anal fin rays are numerous in the Bathygadidae and Macrouridae, but are seldom reported. Classification sequence and sources of meristic data: Eschmeyer, 1990; Fahay and Markle, 1984; Fahay, 1989; Cohen et al., 1990; Iwamoto, 2002; Iwamoto and Cohen, 2002a; 2002b; Merrett, 2003. PrC = principal caudal rays; ~ = approximately Family Precaudal Total Dorsal Anal Pectoral Pelvic Species Vertebrae Vertebrae Fin Rays Fin Rays Fin Rays Fin Rays Bregmacerotidae Bregmaceros atlanticus 14 53–55 47–56 49–58 16–21 5–7 Bregmaceros cantori 14 45–49 45–49 45–49 16–23 (family) 5–7 Bregmaceros sp. 14–15 52–59 52–59 58–69 16–23 (family) 5–7 Bregmaceros houdei 13–14 47–50 47–50 41–46 16–23 (family) 5–7 Family Precaudal Total First + Second Anal Pectoral Pelvic Species Vertebrae Vertebrae Dorsal Fin Rays Fin Rays Fin Rays Fin Rays Bathygadidae Bathygadus favosus 12–14 ~70 9–11+125 110 15–18 9(10) Gadomus dispar 12–13 80+ 12–13 – 18–20 8 Gadomus longifilis 11–13 – 9–11 – 14–16 8–9 Macrouridae Caelorinchus caribbeus 11–12 – 11–12+>110 >110 17–20 7 Caelorinchus coelorhynchus 11–12 – 10–11 – (17)18–20(21) 7 Caelorinchus occa 12–13 – 9–11 – 17–20 7 Coryphaenoides alateralis – 13 – 21–23 8 Coryphaenoides armatus 13–15 – 10–12+~125 ~135 19–21 10–11 Coryphaenoides brevibarbis 12–13 – 9 – 19–20 8–9 Coryphaenoides carapinus 12–15 – 10–11+100 117 17–20 9–11 Coryphaenoides guentheri -

The Ecology of Arctic Cod (Boreogadus Saida) and Interactions with Seabirds, Seals, and Whales in the Canadian Arctic
The Ecology of Arctic Cod (Boreogadus saida) and Interactions with Seabirds, Seals, and Whales in the Canadian Arctic by Jordan K. Matley A Thesis submitted to the Faculty of Graduate Studies of The University of Manitoba in partial fulfilment of the requirements of the degree of MASTER OF SCIENCE Department of Biological Sciences University of Manitoba Winnipeg, Manitoba Copyright © 2012 by Jordan K. Matley i Abstract This thesis investigates the foraging behaviour of Arctic cod (Boreogadus saida) and its predators during the summer in Allen Bay and Resolute Bay, Cornwallis Island, Nunavut, Canada. Major findings included the identification of Arctic cod, ringed seal (Pusa hispida), beluga (Delphinapterus leucas), and narwhal (Monodon monoceros) diet shifts in response to seasonal prey availability; calculation of isotopic diet-tissue discrimination factors for Arctic cod, ringed seals, and whales based on local tissue and stomach content sampling; and determination of cues that predators use to optimize foraging, such as the presence of schools. Additionally, I quantified seabird feeding and interspecific interactions such kleptoparasitism and found that black-legged kittiwakes (Rissa tridactyla) and northern fulmars (Fulmarus glacialis) captured cod directly but lost many to parasitic jaegers (Stercorarius parasiticus) and glaucous gulls (Larus hyperboreus). Finally, I determined that schools of cod were important prey sources for northern fulmars, glaucous gulls, and whales however non-schooling cod were a significant source for black-legged kittiwakes and ringed seals. ii Acknowledgements Numerous people and funding sources are responsible for the completion of this thesis. First, I thank my parents for being supportive of all my endeavours. Second, I thank staff, professors, and students in the Department of Biological Sciences at the University of Manitoba. -

Descriptive Key to the Otoliths of Gadid Fishes of the Bering, Chukchi, and Beaufort Seas KATHRYN J
ARCTIC VOL. 34, NO. 1 (MARCH 1981), P. 55-59 Descriptive Key to the Otoliths of Gadid Fishes of the Bering, Chukchi, and Beaufort Seas KATHRYN J. FROST’ ABSTRACT. An illustrated key with supplementary descriptive material is presented for six species or species groups of gadid fishes which are of trophic importance in the Bering, Chukchi, and Beaufort seas. These species include: Arcrogadus spp. Djagin, Boreogadus saida (Lepechin), Eleginus gracilis (Tilesius), Gadus macrocephalus Tilesius, Microgadus proximus (Girard), and Theragra chalcogramma (Pallas). RESUME. Une clC d’identificationillustree par des figures avec un complement descriptif est ici presentee pour six espbces ou groupes d’espbces de poissons de la famiile des gadidCs, lesquels ont une importance au point de vue trophique dans les mers de BCring, des Tchouktches et deBeaufort. Ces espbcescomprennent: Arctogadus spp. Djagin, Boreogadus saida (Lepechin), Eleginus gracilis (Tilesius), Gadus macrocephalus Tilesius, Microgadus proximus (Girard), et Theragra chalcogramma (Pallas). Traduit par Jean-Guy Brossard, Laboratoire d’Archeologie de I’Universit6 due Quebec A Montreal. Key words: otolith, gadid fishes, Arctogadus, Boreogadus, Eleginus, Gadus, Microgadus, Theragra INTRODUCTION size or when certain features vary such that an otolith of Investigations of food habits of marine animals almost one species closelyresembles that of other species. Furth- invariably involve analysisof stomach contents. Success- er, keys,are often usedby readers who have little familiar- ful stomach contents analysis usually requires that prey ity with otoliths and limited access to comparative mate- items be recognized by characteristic fragments. In this rial, and who therefore require more detailed descriptive respect the sagittal otoliths of bony fishes are very useful material. -

Arctic Cod (Boreogadus Saida) As Prey: Fish Length-Energetics Relationships in the Beaufort Sea and Hudson Bay B
ARCTIC VOL. 66, NO. 2 (JUNE 2013) P. 191 – 196 Arctic Cod (Boreogadus saida) as Prey: Fish Length-Energetics Relationships in the Beaufort Sea and Hudson Bay B. BRITTEN HARTER,1,2 KYLE H. ELLIOTT,1 GEORGE J. DIVOKY3 and GAIL K. DAVOREN1 (Received 1 May 2012; accepted in revised form 13 November 2012) ABSTRACT. Although Arctic cod (Boreogadus saida) is widely recognized as an important trophic link to top predators in Arctic marine ecosystems, the challenges of conducting fieldwork in the Arctic make this species difficult to study. We establish some basic relationships to improve prey energetics modeling when only in-field parameters (e.g., fork length) can be measured. We investigated the intraspecific relationships among energy density, fork length, mass, and water content for Arctic cod captured by Black Guillemots and Thick-billed Murres at two sites (Western Beaufort and Hudson Bay). Dry -1 energy density was similar between sites (21.6 – 22.2 kJ g ) and increased with fork length (Dry EDkJ/g = 0.028 (± 0.01) • Fork Lengthmm + 18.12 (± 1.33). Even though fish lost some water as they were transported to the nest by avian predators, wet energy density also increased with fork length. We suggest that environmental conditions had a similar effect on growth at these two locations although they occur in very different oceanographic regimes. Arctic cod, especially large cod, is one of the most energy-rich prey species in the Arctic. Our results highlight the importance of this valuable prey to Arctic ecosystems and the utility of using seabirds opportunistically as samplers of the marine environment.