Omics” Research and Systems Medicine
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biological Organization and Pathology: Three Views on the Normativity of Medicine
Chapter 8 Biological Organization and Pathology: Three Views on the Normativity of Medicine Arantza Etxeberria A b s t r a c t Medical knowledge aims to identify different diseases as wrong conditions of biological organization. One main issue within the fi eld of the phi- losophy of medicine is the question of just how confi dent we can be that what we know about biological organization will help us to identify diseases and propose cures or treatments for them. The concept of biological organization is a complex abstraction which requires the coexistence of constitutive, interactive and experien- tial aspects; while the main attempts at naturalist descriptions of the concept (functional, mechanistic and systemic) fail to be fully comprehensive. Different arguments have supported a naturalist normativity in medicine; the strongest such perspective contrasts the normal or typical state of organizational elements with their “broken” versions. However, the complexity of biological organization sug- gests that there are multiple ways of being healthy or diseased. Thus, the normative goal of medicine of identifying diseases encounters two fundamental questions: (1) Is biology itself normative and can it defi ne the “natural” state? (2) Can medi- cine rely on knowledge other than biological knowledge to identify what goes wrong? As a normative discipline, medicine comes into confl ict with the multiplicity in the very ontology of diseases, which needs to be complemented with epistemic pluralism. Philosophy of medicine therefore needs to explore the sources of that normativity. Keywords Naturalism • Constructivism • Normative and descriptive • Functional approach • Normal–broken framework A. Etxeberria (*) Department of Logic and Philosophy of Science , University of the Basque Country UPV- EHU , Tolosa Hiribidea 70 , 20018 Donostia-San Sebastián , Spain e-mail: [email protected] © Springer International Publishing Switzerland 2016 121 É. -

The Road from Systems Biology to Systems Medicine
nature publishing group Review The road from systems biology to systems medicine Olaf Wolkenhauer1,2, Charles Auffray3, Robert Jaster4, Gustav Steinhoff5 and Olaf Dammann6,7 As research institutions prepare roadmaps for “systems datasets, requiring not only new computational platforms to medicine,” we ask how this differs from applications of systems manage data but most importantly, requiring new ways of biology approaches in medicine and what we (should) have thinking, including the application and development of meth- learned from about one decade of funding in systems biology. odologies from the mathematical sciences; and (iv) to address After surveying the area, we conclude that systems medicine clinical questions with statistical, mathematical, computational, is the logical next step and necessary extension of systems molecular, and cell-biological methodologies requires strategic biology, and we focus on clinically relevant applications. We efforts to motivate and sustain cross-disciplinary collaborations. specifically discuss three related notions. First, more interdis- Applying systems approaches in a clinical setting, practical, ciplinary collaborations are needed to face the challenges of i.e., formal/legal and computational issues of data collection and integrating basic research and clinical practice: integration, sharing are the most immediate challenge and potential threat analysis, and interpretation of clinical and nonclinical data for to progress. Although these issues are crucial and most press- diagnosis, prognosis, and therapy require advanced statistical, ing, we shall here emphasize the role of mathematical modeling computational, and mathematical tools. Second, strategies are as one aspect that is easily forgotten when it comes to setting required to (i) develop and maintain computational platforms priorities. -

Applying Systems Biology to Biomedical Research and Health Care
Schleidgen et al. BMC Health Services Research (2017) 17:761 DOI 10.1186/s12913-017-2688-z RESEARCH ARTICLE Open Access Applying systems biology to biomedical research and health care: a précising definition of systems medicine Sebastian Schleidgen1*† , Sandra Fernau2†, Henrike Fleischer3†, Christoph Schickhardt4†, Ann-Kristin Oßa4 and Eva C. Winkler4 Abstract Background: Systems medicine has become a key word in biomedical research. Although it is often referred to as P4-(predictive, preventive, personalized and participatory)-medicine, it still lacks a clear definition and is open to interpretation. This conceptual lack of clarity complicates the scientific and public discourse on chances, risks and limits of Systems Medicine and may lead to unfounded hopes. Against this background, our goal was to develop a sufficiently precise and widely acceptable definition of Systems Medicine. Methods: In a first step, PubMed was searched using the keyword “systems medicine”. A data extraction tabloid was developed putting forward a means/ends-division. Full-texts of articles containing Systems Medicine in title or abstract were screened for definitions. Definitions were extracted; their semantic elements were assigned as either means or ends. To reduce complexity of the resulting list, summary categories were developed inductively. In a second step, we applied six criteria for adequate definitions (necessity, non-circularity, non-redundancy, consistency, non-vagueness, and coherence) to these categories to derive a so-called précising definition of Systems Medicine. Results: We identified 185 articles containing the term Systems Medicine in title or abstract. 67 contained at least one definition of Systems Medicine. In 98 definitions, we found 114 means and 132 ends. -

Can Systems Medicine Integrate Scientific and Humanistic
View metadata, citation and similar papers at core.ac.uk brought to you by CORE bs_bs_banner provided by NORA - Norwegian Open Research Archives Journal of Evaluation in Clinical Practice ISSN 1365-2753 Getting personal: can systems medicine integrate scientific and humanistic conceptions of the patient? Henrik Vogt MD,1 Elling Ulvestad MD Dr Med,3,4 Thor Eirik Eriksen Cand Polit5,6 and Linn Getz MD PhD2 1PhD Candidate, 2Professor, General Practice Research Unit, Department of Public Health and General Practice, Norwegian University of Science and Technology (NTNU), Trondheim, Norway 3Professor, Department of Microbiology, The Gade Institute, Haukeland University Hospital, Bergen, Norway 4Professor, Department of Clinical Science, University of Bergen, Bergen, Norway 5PhD Candidate, Department of Work and Environmental Medicine, Hospital of North Norway, Tromsø, Norway 6Senior Advisor, Faculty of Humanities, Social Sciences and Education, Department of Philosophy, UiT – The Arctic University of Norway, Tromsø, Norway Keywords Abstract agency, biopsychosocial medicine, downward causation, emergence, Rationale, aims and objectives The practicing doctor, and most obviously the primary experience, free will, general practice, care clinician who encounters the full complexity of patients, faces several fundamental but generalism, holism, humanistic medicine, intrinsically related theoretical and practical challenges – strongly actualized by so-called intentionality, medically unexplained medically unexplained symptoms (MUS) and multi-morbidity. Systems medicine, which is symptoms, mind-body problem, the emerging application of systems biology to medicine and a merger of molecular multi-morbidity, narrative medicine, biomedicine, systems theory and mathematical modelling, has recently been proposed as a ontology, patient-centred medicine, primary care-centered strategy for medicine that promises to meet these challenges. -

From Systems Biology to P4 Medicine: Applications in Respiratory Medicine
SERIES PERSONALISED MEDICINE From systems biology to P4 medicine: applications in respiratory medicine Guillaume Noell1,2, Rosa Faner1,2 and Alvar Agustí1,2,3 Number 4 in the Series “Personalised medicine in respiratory diseases” Edited by Renaud Louis and Nicolas Roche Affiliations: 1Institut d’Investigacions Biomediques August Pi i Sunyer (IDIBAPS), Barcelona, Spain. 2CIBER Enfermedades Respiratorias (CIBERES), Barcelona, Spain. 3Respiratory Institute, Hospital Clinic, Universitat de Barcelona, Barcelona, Spain. Correspondence: Alvar Agustí, Respiratory Institute, Hospital Clínic, Villarroel 170, 08036 Barcelona, Spain. E-mail: [email protected] @ERSpublications Systems biology and network medicine have the potential to transform medical research and practice http://ow.ly/r3jR30hf35x Cite this article as: Noell G, Faner R, Agustí A. From systems biology to P4 medicine: applications in respiratory medicine. Eur Respir Rev 2018; 27: 170110 [https://doi.org/10.1183/16000617.0110-2017]. ABSTRACT Human health and disease are emergent properties of a complex, nonlinear, dynamic multilevel biological system: the human body. Systems biology is a comprehensive research strategy that has the potential to understand these emergent properties holistically. It stems from advancements in medical diagnostics, “omics” data and bioinformatic computing power. It paves the way forward towards “P4 medicine” (predictive, preventive, personalised and participatory), which seeks to better intervene preventively to preserve health or therapeutically to cure -

Artificial Intelligence: a Disruptive Tool for a Smarter Medicine
European Review for Medical and Pharmacological Sciences 2020; 24: 7462-7474 Artificial intelligence: a disruptive tool for a smarter medicine C.M. GALMARINI, M. LUCIUS Topazium Artificial Intelligence, Madrid, Spain Abstract. – OBJECTIVE: Although highly dinary: in just 100 years, life expectancy has successful, the medical R&D model is failing rocketed from approximately 45 to 72 years. at improving people’s health due to a series of However, there is still a lot of work to be done flaws and defects inherent to the model itself. since the figure for all-cause mortality stands at A new collective intelligence, incorporating hu- almost 57 million. This figure includes 47 mil- man and artificial intelligence (AI) could over- come these obstacles. Because AI will play a lions of adults with chronic, non-communicable key role in this new collective intelligence, it diseases, and 6.5 millions of children, of whom is necessary that those involved in healthcare 5.6 million are under the age of five1. In spite of have a general knowledge of how these technol- the massive economic efforts in R&D and inno- ogies work. With this comprehensive review, we vation, global health improvement seems to have intend to provide it. MATERIALS AND METHODS: reached a plateau. The current model of medical A broad-rang- research is showing difficulties due to a series of ing search has been undertaken on institutional 2 and non-institutional websites in order to identi- limitations . Firstly, its current framework mir- fy relevant papers, comments and reports. rors a linear and sequential process, which has RESULTS: We firstly describe the flaws and been the only paradigm accepted so far. -

Rethinking the Pragmatic Systems Biology and Systems-Theoretical Biology Divide: Toward a Complexity-Inspired Epistemology of Systems Biomedicine T
Medical Hypotheses 131 (2019) 109316 Contents lists available at ScienceDirect Medical Hypotheses journal homepage: www.elsevier.com/locate/mehy Rethinking the pragmatic systems biology and systems-theoretical biology divide: Toward a complexity-inspired epistemology of systems biomedicine T Srdjan Kesić Department of Neurophysiology, Institute for Biological Research “Siniša Stanković”, University of Belgrade, Despot Stefan Blvd. 142, 11060 Belgrade, Serbia ARTICLE INFO ABSTRACT Keywords: This paper examines some methodological and epistemological issues underlying the ongoing “artificial” divide Systems biology between pragmatic-systems biology and systems-theoretical biology. The pragmatic systems view of biology has Cybernetics encountered problems and constraints on its explanatory power because pragmatic systems biologists still tend Second-order cybernetics to view systems as mere collections of parts, not as “emergent realities” produced by adaptive interactions Complexity between the constituting components. As such, they are incapable of characterizing the higher-level biological Complex biological systems phenomena adequately. The attempts of systems-theoretical biologists to explain these “emergent realities” Epistemology of complexity using mathematics also fail to produce satisfactory results. Given the increasing strategic importance of systems biology, both from theoretical and research perspectives, we suggest that additional epistemological and methodological insights into the possibility of further integration between -

Systems Approaches to Biology and Disease Enable Translational Systems Medicine
Available online at www.sciencedirect.com GENOMICS PROTEOMICS & BIOINFORMATICS Genomics Proteomics Bioinformatics 10 (2012) 181–185 www.elsevier.com/locate/gpb Review Systems Approaches to Biology and Disease Enable Translational Systems Medicine Leroy Hood ⇑, Qiang Tian ⇑ Institute for Systems Biology, Seattle, WA 98109-5234, USA Received 31 July 2012; revised 9 August 2012; accepted 14 August 2012 Available online 23 August 2012 Abstract The development and application of systems strategies to biology and disease are transforming medical research and clinical practice in an unprecedented rate. In the foreseeable future, clinicians, medical researchers, and ultimately the consumers and patients will be increasingly equipped with a deluge of personal health information, e.g., whole genome sequences, molecular profiling of diseased tissues, and periodic multi-analyte blood testing of biomarker panels for disease and wellness. The convergence of these practices will enable accurate prediction of disease susceptibility and early diagnosis for actionable preventive schema and personalized treatment regimes tailored to each individual. It will also entail proactive participation from all major stakeholders in the health care system. We are at the dawn of predictive, preventive, personalized, and participatory (P4) medicine, the fully implementation of which requires marrying basic and clinical researches through advanced systems thinking and the employment of high-throughput technologies in genomics, pro- teomics, nanofluidics, single-cell analysis, -

Past Seminars (2011- 2019)
Division of Oncology Biostatistics and Bioinformatics Research Program in Quantitative Sciences Past Seminars (2011- 2019) May 30, 2019 "Simultaneous Testing Procedure for Mediation Effect of DNA Methylation in Genome-Wide Epigenetic Studies" Jincheng Shen, PhD Adjunct Assistant Professor, Internal Medicine/Family and Preventive Medicine Assistant Professor, Population Health Sciences University of Utah Salt Lake City, UT May 2, 2019 "Health disparities calculator (HD*Calc): a tool for estimating health disparities using public health surveillance data" Mandi Yu, PhD Associate Professor Statistical Research and Applications Branch National Cancer Institute Bethesda, MD April 18, 2019 CANCELLED March 21, 2019 "A Bayesian Nonparametric Model for Classification of Longitudinal Profiles" Jeremy Gaskins, PhD Assistant Professor Department of Bioinformatics and Biostatistics School of Public Health and Information Sciences University of Louisville Louisville, KY February 21, 2019 "Study Design and Analysis Considerations in CBER Innovative Biologic Products" Min (Annie) Lin, PhD Mathematical Statistician Division of Biostatistics FDA/CBER/OBE Silver Spring, MD January 17, 2019 "Integrative Computational Approaches to the Understanding and Treatment of Cancer" Jill Mesirov, PhD Associated Vice Chancellor for Computational Health Sciences Professor, Department of Medicine University of California - San Diego La Jolla, CA 2018 December 6, 2018 "Discovery, Design and Analysis of Multidrug Combinations: Nonclinical Modeling to Clinical Trials" Ming -

From Data Gathering to Systems Medicine
Cardiovascular Research (2013) 97, 599–600 EDITORIAL doi:10.1093/cvr/cvt017 From data gathering to systems medicine Manuel Mayr* King’s British Heart Foundation Centre, King’s College London, 125 Coldharbour Lane, London SE5 9NU, UK Online publish-ahead-of-print 5 February 2013 While molecular interactions have been a research focus for many the quantification of transcript levels that do not necessarily corres- 9 years, the advent of updated molecular profiling methods has pond to protein abundance. Downloaded from shifted the attention towards a more integrative approach. The While ‘-omics’ technologies are widely utilized for data gathering, ‘-omics’ technologies—genomics, transcriptomics, proteomics, and interpreting this overwhelming amount of data represents a major metabolomics—allow us to gather a vast amount of information at hurdle. Two reviews are dedicated to this topic: Quinn et al.10 the level of the genome, transcriptome, proteome, and metabolome. describes how combining the wet laboratory and the dry laboratory However, these updated technologies have also brought about the may alleviate the current bottleneck with single cell, tissue, and http://cardiovascres.oxfordjournals.org/ challenge to understand the complex interplay of molecular changes whole-heart studies of cardiac electrical and mechanical function related to cardiovascular disease. This will be the formidable task of and stresses that any computational model will depend on the input systems biology.1,2 In this thematic Mini-Spotlight issue, four reviews of high-quality data. Azuaje et al.11 explores the use of computational summarize strategies how to advance this field. methods to predict drug interactions. Just like pathogenetic mechan- Classic genomics aims to link variations in the DNA sequence isms, drug effects are also looked at in a reductionist fashion, which directly to distinct phenotypes. -
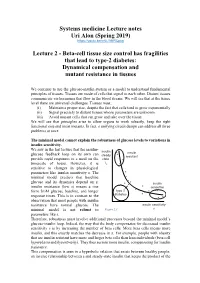
Lecture 2 - Beta-Cell Tissue Size Control Has Fragilities That Lead to Type-2 Diabetes: Dynamical Compensation and Mutant Resistance in Tissues
Systems medicine Lecture notes Uri Alon (Spring 2019) https://youtu.be/m6LYiNRCpxg Lecture 2 - Beta-cell tissue size control has fragilities that lead to type-2 diabetes: Dynamical compensation and mutant resistance in tissues We continue to use the glucose-insulin system as a model to understand fundamental principles of tissues. Tissues are made of cells that signal to each other. Distant tissues communicate via hormones that flow in the blood stream. We will see that at the tissue level there are universal challenges. Tissues must: (i) Maintain a proper size, despite the fact that cells tend to grow exponentially (ii) Signal precisely to distant tissues whose parameters are unknown. (iii) Avoid mutant cells that can grow and take over the tissue. We will see that principles arise to allow organs to work robustly, keep the right functional size and resist mutants. In fact, a unifying circuit design can address all three problems at once. The minimal model cannot explain the robustness of glucose levels to variations in insulin sensitivity. We saw in the last lecture that the insulin- insulin glucose feedback loop on its own can insulin steady resistant provide rapid responses to a meal on the state timescale of hours. However, it is Ist sensitive to changes in physiological parameters like insulin sensitivity s. The minimal model predicts that baseline glucose and its dynamics depend on s: insulin insulin resistance (low s) means a rise sensetive form 5mM glucose baseline, and longer type 2 response times. This is in contrast to the diabetes observation that most people with insulin resistance have normal glucose. -

Systems Biology and Systems Medicine
Systems Biology and Systems Medicine Lee Hood Institute for Systems Biology, Seattle Biology is an Informational Science There are two types of Biological Information • The digital information of the genome • The environmental information that impinges upon and modifies the digital information. Two Types of Systems Biology • Decipher function of molecular machines-- execute biological functions • Decipher function of biological networks-- capture, transmit, integrate and disperse biological information • Two types of biological networks: protein networks and gene regulatory networks Most Sophisticated Integrated Biological Network Defined to Date Sea Urchin Larval Development Most Sophisticated Integrated Biological Network Defined to Date Networks Process Biological Information: Transmit, Integrate, Execute Hierarchical or Multiscalar Levels of Biological Information DNA Top down, bottom up and mRNA both ways from middle Protein Protein interactions and biomodules Protein and gene networks Cells Organs Level of System Analysis Individuals Populations Connect to Digital Core Ecologies Integration of Different Levels Systems Biology at ISB Technology Has Transformed Contemporary Systems Biology Six essential features of contemporary systems biology Quantitative measurements for all types of biological information. Global measurements--measure dynamic changes in all genes, mRNAs, proteins, etc, across state changes. Computational and mathematically integrate different data types--DNA, RNA, Protein, Interactions, etc.--to capture distinct