Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cytochrome P450 Enzymes in Oxygenation of Prostaglandin Endoperoxides and Arachidonic Acid
Comprehensive Summaries of Uppsala Dissertations from the Faculty of Pharmacy 231 _____________________________ _____________________________ Cytochrome P450 Enzymes in Oxygenation of Prostaglandin Endoperoxides and Arachidonic Acid Cloning, Expression and Catalytic Properties of CYP4F8 and CYP4F21 BY JOHAN BYLUND ACTA UNIVERSITATIS UPSALIENSIS UPPSALA 2000 Dissertation for the Degree of Doctor of Philosophy (Faculty of Pharmacy) in Pharmaceutical Pharmacology presented at Uppsala University in 2000 ABSTRACT Bylund, J. 2000. Cytochrome P450 Enzymes in Oxygenation of Prostaglandin Endoperoxides and Arachidonic Acid: Cloning, Expression and Catalytic Properties of CYP4F8 and CYP4F21. Acta Universitatis Upsaliensis. Comprehensive Summaries of Uppsala Dissertations from Faculty of Pharmacy 231 50 pp. Uppsala. ISBN 91-554-4784-8. Cytochrome P450 (P450 or CYP) is an enzyme system involved in the oxygenation of a wide range of endogenous compounds as well as foreign chemicals and drugs. This thesis describes investigations of P450-catalyzed oxygenation of prostaglandins, linoleic and arachidonic acids. The formation of bisallylic hydroxy metabolites of linoleic and arachidonic acids was studied with human recombinant P450s and with human liver microsomes. Several P450 enzymes catalyzed the formation of bisallylic hydroxy metabolites. Inhibition studies and stereochemical analysis of metabolites suggest that the enzyme CYP1A2 may contribute to the biosynthesis of bisallylic hydroxy fatty acid metabolites in adult human liver microsomes. 19R-Hydroxy-PGE and 20-hydroxy-PGE are major components of human and ovine semen, respectively. They are formed in the seminal vesicles, but the mechanism of their biosynthesis is unknown. Reverse transcription-polymerase chain reaction using degenerate primers for mammalian CYP4 family genes, revealed expression of two novel P450 genes in human and ovine seminal vesicles. -

Synonymous Single Nucleotide Polymorphisms in Human Cytochrome
DMD Fast Forward. Published on February 9, 2009 as doi:10.1124/dmd.108.026047 DMD #26047 TITLE PAGE: A BIOINFORMATICS APPROACH FOR THE PHENOTYPE PREDICTION OF NON- SYNONYMOUS SINGLE NUCLEOTIDE POLYMORPHISMS IN HUMAN CYTOCHROME P450S LIN-LIN WANG, YONG LI, SHU-FENG ZHOU Department of Nutrition and Food Hygiene, School of Public Health, Peking University, Beijing 100191, P. R. China (LL Wang & Y Li) Discipline of Chinese Medicine, School of Health Sciences, RMIT University, Bundoora, Victoria 3083, Australia (LL Wang & SF Zhou). 1 Copyright 2009 by the American Society for Pharmacology and Experimental Therapeutics. DMD #26047 RUNNING TITLE PAGE: a) Running title: Prediction of phenotype of human CYPs. b) Author for correspondence: A/Prof. Shu-Feng Zhou, MD, PhD Discipline of Chinese Medicine, School of Health Sciences, RMIT University, WHO Collaborating Center for Traditional Medicine, Bundoora, Victoria 3083, Australia. Tel: + 61 3 9925 7794; fax: +61 3 9925 7178. Email: [email protected] c) Number of text pages: 21 Number of tables: 10 Number of figures: 2 Number of references: 40 Number of words in Abstract: 249 Number of words in Introduction: 749 Number of words in Discussion: 1459 d) Non-standard abbreviations: CYP, cytochrome P450; nsSNP, non-synonymous single nucleotide polymorphism. 2 DMD #26047 ABSTRACT Non-synonymous single nucleotide polymorphisms (nsSNPs) in coding regions that can lead to amino acid changes may cause alteration of protein function and account for susceptivity to disease. Identification of deleterious nsSNPs from tolerant nsSNPs is important for characterizing the genetic basis of human disease, assessing individual susceptibility to disease, understanding the pathogenesis of disease, identifying molecular targets for drug treatment and conducting individualized pharmacotherapy. -

Essential Role of the Cytochrome P450 CYP4F22 in the Production of Acylceramide, the Key Lipid for Skin Permeability Barrier Formation
Essential role of the cytochrome P450 CYP4F22 in the production of acylceramide, the key lipid for skin permeability barrier formation Yusuke Ohnoa, Shota Nakamichia, Aya Ohkunia, Nozomi Kamiyamaa, Ayano Naoeb, Hisashi Tsujimurab, Urara Yokoseb, Kazumitsu Sugiurac, Junko Ishikawab, Masashi Akiyamac, and Akio Kiharaa,1 aLaboratory of Biochemistry, Faculty of Pharmaceutical Sciences, Hokkaido University, Kita-ku, Sapporo 060-0812, Japan; bKao Corporation, Haga-gun, Tochigi 321-3497, Japan; and cDepartment of Dermatology, Nagoya University Graduate School of Medicine, Showa-ku, Nagoya 466-8550, Japan Edited by David W. Russell, University of Texas Southwestern Medical Center, Dallas, TX, and approved May 21, 2015 (received for review February 19, 2015) A skin permeability barrier is essential for terrestrial animals, and its lamellae and/or to stabilize the multiple lipid layers. Linoleic impairment causes several cutaneous disorders such as ichthyosis and acid is one of the essential FAs, and its deficiency causes ich- atopic dermatitis. Although acylceramide is an important lipid for the thyosis symptoms resulting from a failure to form normal acyl- skin permeability barrier, details of its production have yet to be de- ceramide (8). Ichthyosis is a cutaneous disorder accompanied termined, leaving the molecular mechanism of skin permeability bar- by dry, thickened, and scaly skin; it is caused by a barrier ab- rier formation unclear. Here we identified the cytochrome P450 gene normality. In patients who have atopic dermatitis, both total CYP4F22 (cytochrome P450, family 4, subfamily F, polypeptide 22) as ceramide levels and the chain length of ceramides are de- the long-sought fatty acid ω-hydroxylase gene required for acylcer- creased, and ceramide composition is altered also (9–11). -

Robert Foti to Cite This Version
Characterization of xenobiotic substrates and inhibitors of CYP26A1, CYP26B1 and CYP26C1 using computational modeling and in vitro analyses Robert Foti To cite this version: Robert Foti. Characterization of xenobiotic substrates and inhibitors of CYP26A1, CYP26B1 and CYP26C1 using computational modeling and in vitro analyses. Agricultural sciences. Université Nice Sophia Antipolis, 2016. English. NNT : 2016NICE4033. tel-01376678 HAL Id: tel-01376678 https://tel.archives-ouvertes.fr/tel-01376678 Submitted on 5 Oct 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Université de Nice-Sophia Antipolis Thèse pour obtenir le grade de DOCTEUR DE L’UNIVERSITE NICE SOPHIA ANTIPOLIS Spécialité : Interactions Moléculaires et Cellulaires Ecole Doctorale : Sciences de la Vie et de la Santé (SVS) Caractérisation des substrats xénobiotiques et des inhibiteurs des cytochromes CYP26A1, CYP26B1 et CYP26C1 par modélisation moléculaire et études in vitro présentée et soutenue publiquement par Robert S. Foti Le 4 Juillet 2016 Membres du jury Dr. Danièle Werck-Reichhart Rapporteur Dr. Philippe Roche Rapporteur Pr. Serge Antonczak Examinateur Dr. Philippe Breton Examinateur Pr. Philippe Diaz Examinateur Dr. Dominique Douguet Directrice de thèse 1 1. Table of Contents 1. Table of Contents .............................................................................................................................. -

Metabolic Activation and Toxicological Evaluation of Polychlorinated Biphenyls in Drosophila Melanogaster T
www.nature.com/scientificreports OPEN Metabolic activation and toxicological evaluation of polychlorinated biphenyls in Drosophila melanogaster T. Idda1,7, C. Bonas1,7, J. Hofmann1, J. Bertram1, N. Quinete1,2, T. Schettgen1, K. Fietkau3, A. Esser1, M. B. Stope4, M. M. Leijs3, J. M. Baron3, T. Kraus1, A. Voigt5,6 & P. Ziegler1* Degradation of polychlorinated biphenyls (PCBs) is initiated by cytochrome P450 (CYP) enzymes and includes PCB oxidation to OH-metabolites, which often display a higher toxicity than their parental compounds. In search of an animal model refecting PCB metabolism and toxicity, we tested Drosophila melanogaster, a well-known model system for genetics and human disease. Feeding Drosophila with lower chlorinated (LC) PCB congeners 28, 52 or 101 resulted in the detection of a human-like pattern of respective OH-metabolites in fy lysates. Feeding fies high PCB 28 concentrations caused lethality. Thus we silenced selected CYPs via RNA interference and analyzed the efect on PCB 28-derived metabolite formation by assaying 3-OH-2′,4,4′-trichlorobiphenyl (3-OHCB 28) and 3′-OH-4′,4,6′-trichlorobiphenyl (3′-OHCB 28) in fy lysates. We identifed several drosophila CYPs (dCYPs) whose knockdown reduced PCB 28-derived OH-metabolites and suppressed PCB 28 induced lethality including dCYP1A2. Following in vitro analysis using a liver-like CYP-cocktail, containing human orthologues of dCYP1A2, we confrm human CYP1A2 as a PCB 28 metabolizing enzyme. PCB 28-induced mortality in fies was accompanied by locomotor impairment, a common phenotype of neurodegenerative disorders. Along this line, we show PCB 28-initiated caspase activation in diferentiated fy neurons. -

Effect of Genetic Variability in the CYP4F2, CYP4F11 and CYP4F12 Genes on Liver Mrna Levels and Warfarin Response
Effect of genetic variability in the CYP4F2, CYP4F11 and CYP4F12 genes on liver mRNA levels and warfarin response J. Eunice Zhang1, Kathrin Klein2, Andrea L. Jorgensen3, Ben Francis3, Ana Alfirevic1, Stephane Bourgeois4, Panagiotis Deloukas4,5,6, Ulrich M. Zanger2 and Munir Pirmohamed1* 1Wolfson Centre for Personalized Medicine, Department of Molecular & Clinical Pharmacology, The University of Liverpool, UK 2Dr Margarete Fischer-Bosch Institute of Clinical Pharmacology, Stuttgart, and University of Tuebingen, Tuebingen, Germany. 3Department of Biostatistics, The University of Liverpool, UK 4William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, UK 5Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK. 6Princess Al-Jawhara Al-Brahim Centre of Excellence in Research of Hereditary Disorders, King Abdulaziz University, Jeddah, Saudi Arabia. Correspondence: Professor Sir Munir Pirmohamed Wolfson Centre for Personalised Medicine Department of Molecular & Clinical Pharmacology Block A: Waterhouse Buildings The University of Liverpool 1-5 Brownlow Street L69 3GL, UK [email protected] Running Title: Genotype-phenotype assessment of CYP4F12-CYP4F2-CYP4F11 region Keywords: warfarin, pharmacogenetics, mRNA expression, CYP4F2, CYP4F11, CYP4F12 1 Abstract Genetic polymorphisms in the gene encoding cytochrome P450 (CYP) 4F2, a vitamin K oxidase, affect stable warfarin dose requirements and time to therapeutic INR. CYP4F2 is part of the CYP4F gene cluster, which is highly polymorphic and exhibits a high degree of linkage disequilibrium, making it difficult to define causal variants. Our objective was to examine the effect of genetic variability in the CYP4F gene cluster on expression of the individual CYP4F genes and warfarin response. mRNA levels of the CYP4F gene cluster were quantified in human liver samples (n=149) obtained from a well characterized liver bank and fine mapping of the CYP4F gene cluster encompassing CYP4F2, CYP4F11 and CYP4F12 was performed. -

Metabolic Activation of 4-Ipomeanol by Complementary DNA-Expressed Human Cytochromes P-450: Evidence for Species-Specific Metabolism Maciej Czerwinski, Theodore L
[CANCER RESEARCH 51. 4636-4638, September I, 1991] Metabolic Activation of 4-Ipomeanol by Complementary DNA-expressed Human Cytochromes P-450: Evidence for Species-specific Metabolism Maciej Czerwinski, Theodore L. McLemore,1 Richard M. Philpot, Patson T. Nhamburo, Kenneth Korzekwa, Harry V. Gelboin, and Frank J. Gonzalez Laboratory of Molecular Carcinogenesis, Division of Cancer Etiology [M. C., P. T. N., K. K., H. V. G., F. J. G.¡,and Developmental Therapeutics Program, Division of Cancer Treatment [T. L. M.], National Cancer Institute, NIH, Bethesda, Maryland 20892, and Laboratory of Pharmacology, National Institutes of Environmental Health Sciences, NIH, Research Triangle Park, North Carolina 27709 ¡R.M. P.] ABSTRACT Specific enzymes present in human lung that are capable of activating 4-ipomeanol have not been established previously. 4-Ipomeanol is a pulmonary toxin in cattle and rodents that is meta- We therefore studied the ability of 14 P-450s to activate 4- bolically activated by cytochromes P-450 (P-450s). P-450-mediated ac ipomeanol to DNA-binding metabolites using vaccinia virus- tivation of 4-ipomeanol to DNA binding metabolites was evaluated using mediated cDNA expression of P-450s in human Hep G2 cells a vaccinia virus complementary DNA expression system and an in situ DNA-binding assay. Twelve human P-450s and two rodent P-450s were and in situ binding to cellular DNA. We report that several P- expressed in human hepatoma Hep G2 cells and examined for their 450s can catalyze some activation of 4-ipomeanol and at least abilities to metabolically activate this toxin. Three forms, designated three distinct human P-450 forms are capable of high rates of CYP1A2, CYP3A3, and CYP3A4, were able to catalyze significant biotransformation of this compound. -

Recent Advances in P450 Research
The Pharmacogenomics Journal (2001) 1, 178–186 2001 Nature Publishing Group All rights reserved 1470-269X/01 $15.00 www.nature.com/tpj REVIEW Recent advances in P450 research JL Raucy1,2 ABSTRACT SW Allen1,2 P450 enzymes comprise a superfamily of heme-containing proteins that cata- lyze oxidative metabolism of structurally diverse chemicals. Over the past few 1La Jolla Institute for Molecular Medicine, San years, there has been significant progress in P450 research on many fronts Diego, CA 92121, USA; 2Puracyp Inc, San and the information gained is currently being applied to both drug develop- Diego, CA 92121, USA ment and clinical practice. Recently, a major accomplishment occurred when the structure of a mammalian P450 was determined by crystallography. Correspondence: Results from these studies will have a major impact on understanding struc- JL Raucy,La Jolla Institute for Molecular Medicine,4570 Executive Dr,Suite 208, ture-activity relationships of P450 enzymes and promote prediction of drug San Diego,CA 92121,USA interactions. In addition, new technologies have facilitated the identification Tel: +1 858 587 8788 ext 116 of several new P450 alleles. This information will profoundly affect our under- Fax: +1 858 587 6742 E-mail: jraucyȰljimm.org standing of the causes attributed to interindividual variations in drug responses and link these differences to efficacy or toxicity of many thera- peutic agents. Finally, the recent accomplishments towards constructing P450 null animals have afforded determination of the role of these enzymes in toxicity. Moreover, advances have been made towards the construction of humanized transgenic animals and plants. Overall, the outcome of recent developments in the P450 arena will be safer and more efficient drug ther- apies. -
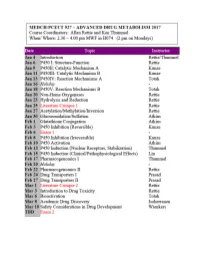
CYTOCHROME P450: Structure-Function
MEDCH 527 AER Jan. 4-6, 2017 CYTOCHROME P450: Structure-Function 1. General P450 Characteristics anD Taxonomy 2. Human P450s – Substrate anD Inhibitor Selectivities 3. Structure-Function Aspects of LiganD BinDing, P450 ReDuction anD Oxygen Activation References P450 Homepage -http:// http://drnelson.uthsc.edu/CytochromeP450.html Nelson DR et al., Pharmacogenetics. 1996 6(1):1-42: Nelson DR et al., Pharmacogenetics. 2004 14(1):1-18. Testa, B. The Biochemistry of Drug Metabolism: A 6 Part Series in Chem. BioDivers. (2006-2008). Sligar, SG. Glimpsing the critical intermediate in cytochrome P450 oxidations. Science. 2010 Nov 12;330(6006):924-5. Johnson EF, et al. Correlating structure and function of drug metabolizing enzymes:Progress and ongoing challenges. Drug Metab. Dispos. 42:9-22 (2014). Zientek MA and Youdim K, Reaction Phenotyping:Advances in experimental strategies used to characterize the contribution of drug metabolizing enzymes. Drug Metab. Dispos. 43:163-181 (2015). Almazroo et al., Drug Metabolism in the Liver. Clin. Liver Dis. 21:1-20 (2017). Absorption, Distribution, Metabolism and Excretion (ADME) of Orally Administered Drugs To reach their sites of action in the body, orally administered drugs must be absorbed from the small intestine, survive first pass metabolism - typically in the liver - before eventually being excreted, usually as drug metabolites in the bile and kidney. Therefore, clinically useful drugs must be able to cross an array of cell membranes, which are composed of a lipid bilayer. Drugs must exhibit an adequate degree of lipophilicity (logP of ~2-4) in order to able to dissolve into this lipoidal environment. Many drug metabolism processes render lipophilic drugs more water-soluble so as to facilitate excretion via the kidneys and bile. -

COVID-19 Pharmacogenetics Lei-Y
medRxiv preprint doi: https://doi.org/10.1101/2020.03.23.20041350; this version posted March 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Genetic Profiles in Pharmacogenes Indicate Personalized Drug Therapy for COVID-19 Running title: COVID-19 pharmacogenetics Lei-Yun Wang1-3#, Jia-Jia Cui1-3#, Qian-Ying OuYang1-3#, Yan Zhan1-3#, Yi-Min Wang7, Xiang-Yang Xu7, Cheng-Xian Guo6*, Ji-Ye Yin1-5* 1Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410078; P. R. China; Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, Changsha 410078; P. R. China. 2Engineering Research Center of Applied Technology of Pharmacogenomics, Ministry of Education, 110 Xiangya Road, Changsha 410078, P. R. China. 3National Clinical Research Center for Geriatric Disorders, 87 Xiangya Road, Changsha 410008, Hunan, P.R. China. 4Hunan Provincial Gynecological Cancer Diagnosis and Treatment Engineering Research Center, Changsha 410078, P. R. China. 5Hunan Key Laboratory of Precise Diagnosis and Treatment of Gastrointestinal Tumor, Changsha 410078, P. R. China. 6Center of Clinical Pharmacology, the Third Xiangya Hospital, Central South University, Changsha, Hunan, 410013, PR China. 7Genetalks Co., Ltd. Building 2, Huxindao, Taiyangshan Road, Qingzhu lake, Changsha 410008, Hunan, P.R. China. #These authors contributed equally to this work. *To whom correspondence should be addressed: Professor Ji-Ye Yin, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008; P. R. China. Tel: +86 731 84805380, Fax: +86 731 82354476, E-mail: [email protected]. -

Urinary Sediment Transcriptomic and Longitudinal Data to Investigate Renal Function Decline in Type 1 Diabetes
ORIGINAL RESEARCH published: 30 April 2020 doi: 10.3389/fendo.2020.00238 Urinary Sediment Transcriptomic and Longitudinal Data to Investigate Renal Function Decline in Type 1 Diabetes Maria Beatriz Monteiro 1, Tatiana S. Pelaes 1, Daniele P. Santos-Bezerra 1, Karina Thieme 2, Antonio M. Lerario 3, Sueli M. Oba-Shinjo 4, Ubiratan F. Machado 2, Marisa Passarelli 5,6, Suely K. N. Marie 4 and Maria Lúcia Corrêa-Giannella 1,6* 1 Laboratório de Carboidratos e Radioimunoensaio (LIM-18), Faculdade de Medicina, Hospital das Clinicas HCFMUSP, Universidade de São Paulo, São Paulo, Brazil, 2 Department of Physiology and Biophysics, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil, 3 Division of Metabolism, Endocrinology, and Diabetes, Department of Internal Medicine, University of Michigan, Ann Arbor, MI, United States, 4 Laboratory of Molecular and Cellular Biology (LIM-15), Faculdade de Medicina, Hospital das Clinicas HCFMUSP, Universidade de São Paulo, São Paulo, Brazil, 5 Laboratório de Lípides (LIM-10), Faculdade de Medicina, Hospital das Clinicas HCFMUSP, Universidade de São Paulo, São Paulo, Brazil, 6 Programa de Pós-graduação em Medicina, Universidade Nove de Julho (UNINOVE), São Paulo, Brazil Edited by: Ondrejˇ Šeda, Introduction: Using a discovery/validation approach we investigated associations Charles University, Czechia between a panel of genes selected from a transcriptomic study and the estimated Reviewed by: glomerular filtration rate (eGFR) decline across time in a cohort of type 1 diabetes Ilse Sofia Daehn, Icahn School of Medicine at Mount (T1D) patients. Sinai, United States Tomislav Bulum, Experimental: Urinary sediment transcriptomic was performed to select highly University of Zagreb, Croatia modulated genes in T1D patients with rapid eGFR decline (decliners) vs. -

Publications
PUBLICATIONS http://www.ncbi.nlm.nih.gov/pubmed/23688132 Nelson CH, Buttrick BR, Isoherranen N. “Therapeutic potential of the inhibition of the retinoic acid hydroxylases CYP26A1 and CYP26B1 by xenobiotics.” Curr Top Med Chem. 2013; 13(12:1402-28. http://www.ncbi.nlm.nih.gov/pubmed/23748241 Parkinson OT, Liggitt HD, Rettie AE, Kelly EJ. “Generalization and characterization of a Cyp4b1 null mouse and the role of CYP4B1 in the activation and toxicity of Ipomeanol.” Toxicol Sci. 2013 Aug; 134(2):243-50. http://www.ncbi.nlm.nih.gov/pubmed/23660171 McDermott CL, Sandmaier BM, Storer B, Li H, Mager DE, Boeckh MJ, Bemer MJ, Knutson J, McCune JS. “Nonrelapse mortality and mycophenolic acid exposure in nonmyeloablative hematopoietic cell transplantation.” Biol Blood Marrow Transplant 2013 Aug; 19(8):1159-66. http://www.ncbi.nlm.nih.gov/pubmed/23620487 Shirasaka Y, Sager JE, Lutz JD, Davis C, Isoherranen N. “Inhibition of CYP2C19 and CYP3A4 by omeprazole metabolites and their contribution to drug-drug interactions.” Drug Metab Dispos. 2013 Jul; 41(7):1414-24. http://www.ncbi.nlm.nih.gov/pubmed/23620487 Shirasaka Y, Sager JE, Lutz JD, Davis C, Isoherranen N. “Inhibition of CYP2C19 and CYP3A4 by omeprazole metabolites and their contribution to drug-drug interactions.” Drug Metabl Dispos. 2013 Jul; 41(7):1414-24. http://www.ncbi.nlm.nih.gov/pubmed/22517924 Guo L, Zhang X, Zhoou D, Okunade AL, Su X. “Stereospecificity of fatty acid 2-hydroxylase and differential functions of 2-hydroxy fatty acid enantiomers.” J Lipid Res. 2013 Jul; 53(7):1327-35. http://www.ncbi.nlm.nih.gov/pubmed/23348398 Roth MY, Nya-Ngatchou JJ, Lin K, Page ST, Anawalt BD, Matsumoto AM, Marck BT, Bremner WJ, Amory JK.