Curriculum Vitae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Annual Report 2008-09
i Contents 1 Executive Director Speaks 1 2 Our Organisation 2 • Mission • Governance • LEAD team 3 Programmes and Progress 4 • The Flagship Programme: Cohort 13 • Voluntary Carbon Market for communities • Sustainable Community Based Tourism: • Youth Summits on Climate Change • Climate Change Ballet “Vasundhara” • Opinion Leader Meet: CSR and Sustainability in India • Climate Change Leaders in the Himalayan Region • Building Sustainable Livelihoods on Tsunami Affected Greater Nicobar Island 4 Thinking Ahead 9 • Flagship Programme Cohort 14 • Environmental Awareness for Youth • Training Programmes on Responsive Climate Governance • Workshops on the Issues and Challenges of Mountain ecosystems • Livelihood Security and Climate Change Adaptation through an Integrated Approach 5 Reaching Out 11 • Networking • Partnerships 6 The Fellows Programme 2008 13 • Fellows Achievements • Fellows Involvement 7 Finance 25 • Auditors Certification • Balance Sheet • Funds Generated 8 Acknowledgement ii Executive Director Speaks Pragya D Varma would have been possible without the support of many of Dear LEADers, you who closely worked with us on these projects. While the annual report 08-09 shall elaborately highlight the specific The last year has seen turbulent times globally and nationally contributions of each bit of support we have got from you all, with the shock waves of the economic crisis being felt around I take this opportunity to make a special mention that LEAD the world and an even more devastating series of terror strikes Fellows have been by us/with us at every step. You have rocking our county, the recent one in Mumbai shaking the helped us in conceiving ideas, formulating project proposals very foundations of our beliefs. -

Sustainable Development and Biodiversity
Sustainable Development and Biodiversity Volume 27 Series Editor Kishan Gopal Ramawat Botany Department, Mohanlal Sukhadia University, Udaipur, India Sustainable Development Goals are best achieved by mechanisms such as research, innovation, and knowledge sharing. This book series aims to help researchers by reporting recent progress and providing complete, comprehensive, and broad subject-based reviews about all aspects of sustainable development and ecological biodiversity. The series explores linkages of biodiversity with delivery of various ecosystem services and it offers a discourse in understanding the biotic and abiotic interactions, ecosystem dynamics, biological invasion, ecological restoration and remediation, diversity of habitats and conservation strategies. It is a broad scoped collection of volumes, addressing relationship between ecosystem processes and biodiversity. It aims to support the global efforts towards achieving sustainable development goals by enriching the scientific literature. The books in the series brings out the latest reading material for botanists, environmentalists, marine biologists, conservationists, policy makers and NGOs working for environment protection. We welcome volumes on the themes – Agroecosystems, Agroforestry, Biodiversity, Biodiversity conservation, Conser- vation of ecosystem, Ecosystem, Endangered species, Forest conservation, Genetic diversity, Global climate change, Hotspots, Impact assessment, Invasive species, Livelihood of people, Plant biotechnology, Plant resource utilization, -
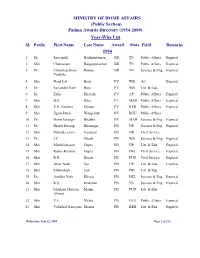
(Public Section) Padma Awards Directory (1954-2009) Year-Wise List Sl
MINISTRY OF HOME AFFAIRS (Public Section) Padma Awards Directory (1954-2009) Year-Wise List Sl. Prefix First Name Last Name Award State Field Remarks 1954 1 Dr. Sarvapalli Radhakrishnan BR TN Public Affairs Expired 2 Shri Chakravarti Rajagopalachari BR TN Public Affairs Expired 3 Dr. Chandrasekhara Raman BR TN Science & Eng. Expired Venkata 4 Shri Nand Lal Bose PV WB Art Expired 5 Dr. Satyendra Nath Bose PV WB Litt. & Edu. 6 Dr. Zakir Hussain PV AP Public Affairs Expired 7 Shri B.G. Kher PV MAH Public Affairs Expired 8 Shri V.K. Krishna Menon PV KER Public Affairs Expired 9 Shri Jigme Dorji Wangchuk PV BHU Public Affairs 10 Dr. Homi Jehangir Bhabha PB MAH Science & Eng. Expired 11 Dr. Shanti Swarup Bhatnagar PB UP Science & Eng. Expired 12 Shri Mahadeva Iyer Ganapati PB OR Civil Service 13 Dr. J.C. Ghosh PB WB Science & Eng. Expired 14 Shri Maithilisharan Gupta PB UP Litt. & Edu. Expired 15 Shri Radha Krishan Gupta PB DEL Civil Service Expired 16 Shri R.R. Handa PB PUN Civil Service Expired 17 Shri Amar Nath Jha PB UP Litt. & Edu. Expired 18 Shri Malihabadi Josh PB DEL Litt. & Edu. 19 Dr. Ajudhia Nath Khosla PB DEL Science & Eng. Expired 20 Shri K.S. Krishnan PB TN Science & Eng. Expired 21 Shri Moulana Hussain Madni PB PUN Litt. & Edu. Ahmed 22 Shri V.L. Mehta PB GUJ Public Affairs Expired 23 Shri Vallathol Narayana Menon PB KER Litt. & Edu. Expired Wednesday, July 22, 2009 Page 1 of 133 Sl. Prefix First Name Last Name Award State Field Remarks 24 Dr. -

And Local Dynamics in Calvary Temple: India’S Fastest Growing Megachurch
Chapter 14 Global, ‘Glocal’ and Local Dynamics in Calvary Temple: India’s Fastest Growing Megachurch Jonathan D. James 1 Introduction Satish Kumar, the founder and senior pastor of Calvary Temple in Andhra Pradesh, India is primarily known for building a gigantic church in 52 days, thus emulating the feat of the Old Testament priest Nehemiah who built the walls of Jerusalem in 52 days (Nehemiah 6: 15). Calvary Temple was built in 2011 to seat 18,000 people (complete with overflow facilities); in 2018 it has close to 200,000 members who worship in four services in the Telugu language and one in English, all on Sunday. In addition to the church, Kumar has a Bible School, conference halls, and other state-of-the-art facilities at the 12-acre property in Hyderabad. In this chapter, I describe how Calvary Temple (hereafter CT) was created, how it operates organisationally, and how the church undertakes its ministry in a nation that is not favourably disposed to Christianity. I also contemplate whether CT is navigating a new path, perhaps as a ‘trailblazer’ for the future of the Christian community in India. Having acquired the status of a megachurch, a phenomenon that emerged in the latter half of the 20th century in the West (especially in the usa) (Thum- ma, Travis and Bird 2005), CT’s size is regarded as an indicator of success. But how did CT grow so big so quickly in a Hindu nation where there were and still are increasing tensions between Hindutva forces and minority religious groups?1 I use the theoretical underpinnings of Robertson (1995) to frame my proposition that CT is a product of both globalisation and glocalisation – the latter being the connection and interplay between the global megachurch movement and the local forces at work in contemporary Indian society. -

Satish Kumar Brief Biography
Satish Kumar brief biography If you are publishing information about Satish in relation to an event, please use one of the following biographies: 100 words Life-long activist and former monk, Satish Kumar has been inspiring global change for over 50 years. In his 20s, Satish undertook a peace-pilgrimage for nuclear disarmament, walking without money from India to America. Now in his 80s, Satish has devoted his life to campaigning for ecological regeneration, social justice, and spiritual fulfilment. An acclaimed author and international speaker, Satish founded The Resurgence Trust, an educational charity that seeks a just future for all. To find more about Satish’s work at The Resurgence Trust and join him in protecting people and planet click here. 200 words Peace-pilgrim, life-long activist and former monk, Satish Kumar has been inspiring global change for over 50 years. Aged 9, Satish renounced the world and joined the wandering Jain monks. Inspired by Gandhi, he decided at 18 that he could achieve more back in the world and soon undertook a peace-pilgrimage, walking without money from India to America in the name of nuclear disarmament. Now in his 80s, Satish has devoted his life to campaigning for ecological regeneration, social justice and spiritual fulfilment. Satish co-founded Schumacher College as well as founding The Resurgence Trust, an educational charity that seeks a just future for all. Satish appears regularly on podcasts, radio and television shows. He has been interviewed by Richard Dawkins, Russell Brand and Annie Lennox, appearing as a guest on Radio 4's Desert Island Discs, Thought for the Day and Midweek. -

Artsand Philanthropy
Arts and Philanthropy Art is a universal language where visual expression evokes raw emotions and is thus an effective medium of conveying the very important message of peace “and harmony. Indeed, powerful images of paintings can leave an everlasting impact on peoples’ minds. Unlike words which are spoken, heard and sometimes forgotten, art on canvas stays forever and so does the message it conveys. — Satish Kumar Modi at the Inaugural Ceremony of the UNESCO Global Forum on the ‘Power of Peace’ ”at Bali, Indonesia. (cover image) — ‘Golden Life’, oil on canvas by Deepak Kumar, student of Final Year, Fine Art, IIFA Inspired by Lord Buddha’s life and teachings, the artist has tried to convey the fact that thoughts based on truthfulness, love and compassion — the ‘golden thoughts’— enable a person to lead a life full of peace, contentment and happiness — a ‘golden life’. (this page) — Collage of experiments — understanding the relation between space and form, Foundation Course, First Year, IIFA. Arts p International Institute of Fine Arts (IIFA) p Art exhibitions p New Arts University, National Capital Region “ p Art show on CNBC p — S.K. Modi Khoj International Artists’ workshop Prof. John Edwards of Charles Wallace Trust demonstrating drawing & colours to Final Year students at IIFA. International Institute of Fine Arts (IIFA) In July 2000 Satish and Abha Modi set up the International Institute of Fine Arts, IIFA, an institute that could nurture young artists, ignite their imagination and motivate them in the field of art and design. Their vision was to promote art and culture and provide quality education in conformity with global standards. -

Civil List of Officers of Indian Postal Service Group -A (As on 01.06.2014)
CIVIL LIST OF OFFICERS OF INDIAN POSTAL SERVICE GROUP -A (AS ON 01.06.2014) DEPARTMENT OF POSTS NEW DELHI DETAILS OF SANCTIONED POSTS IN INDIAN POSTAL SERVICE GROUP ‘A’ GRADE NO. OF POSTS I. MEMBER, POSTAL SERVICES BOARD (HAG +) : 06 II. Sr. DDG/CPMG/CGM (HAG) : 26 III. SENIOR ADMINISTRATIVE GRADE (SAG) : 73 IV. JUNIOR ADMINISTRATIVE GRADE (JAG) (INCLUDING NFSG) : 105 V. SENIOR TIME SCALE (STS) : 198 VI. JUNIOR TIME SCALE (JTS) : 67 VII. RESERVES DEPUTATION RESERVE : 51 TRAINING RESERVE : 30 LEAVE RESERVE : 05 TOTAL 561 02 CIVIL LIST OF OFFICERS OF INDIAN POSTAL SERVICE, GROUP ‘A’ AS ON 01.06.2014 S.N. Name Of The Batch Date of Date of Entry Date of Entry Present Officer (S/Shri) Birth in Service in Grade Deployment GRADE: SECRETARY TO GOVT. OF INDIA/CHAIRPERSON (PSB)/ DG(POSTS) PAY SCALE [Rs. 80000 (FIXED)] 1. Ms. Kavery Banerjee 1978 25.04.56 30.11.78 01.06.14 Secretary (Posts) / Chairperson, PSB / DG (Posts) GRADE: MEMBER, POSTAL SERVICES BOARD PAY SCALE (Rs. 75500 - 80000) 2. Kamleshwar Prasad 1977 02.08.54 07.11.77 15.03.11 Member, PSB 3. Ms. Anjali Devasher 1978 12.07.55 30.11.78 22.05.13 Member, PSB 4. Ms. Kalpana Tewari 1978 13.05.55 30.11.78 09.07.13 Member, PSB 5. Kamlesh Chandra 1978 03.06.54 30.11.78 02.08.13 Member, PSB 6. Shekhar Kumar Sinha 1979 11.07.56 27.12.79 01.12.13 Member, PSB GRADE: CHIEF POSTMASTER GENERAL/SR. DDG [HIGHER ADMINISTRATIVE GRADE (HAG)] PAY SCALE (Rs. -

List of Officers Who Attended Courses at NCRB
List of officers who attened courses at NCRB Sr.No State/Organisation Name Rank YEAR 2000 SQL & RDBMS (INGRES) From 03/04/2000 to 20/04/2000 1 Andhra Pradesh Shri P. GOPALAKRISHNAMURTHY SI 2 Andhra Pradesh Shri P. MURALI KRISHNA INSPECTOR 3 Assam Shri AMULYA KUMAR DEKA SI 4 Delhi Shri SANDEEP KUMAR ASI 5 Gujarat Shri KALPESH DHIRAJLAL BHATT PWSI 6 Gujarat Shri SHRIDHAR NATVARRAO THAKARE PWSI 7 Jammu & Kashmir Shri TAHIR AHMED SI 8 Jammu & Kashmir Shri VIJAY KUMAR SI 9 Maharashtra Shri ABHIMAN SARKAR HEAD CONSTABLE 10 Maharashtra Shri MODAK YASHWANT MOHANIRAJ INSPECTOR 11 Mizoram Shri C. LALCHHUANKIMA ASI 12 Mizoram Shri F. RAMNGHAKLIANA ASI 13 Mizoram Shri MS. LALNUNTHARI HMAR ASI 14 Mizoram Shri R. ROTLUANGA ASI 15 Punjab Shri GURDEV SINGH INSPECTOR 16 Punjab Shri SUKHCHAIN SINGH SI 17 Tamil Nadu Shri JERALD ALEXANDER SI 18 Tamil Nadu Shri S. CHARLES SI 19 Tamil Nadu Shri SMT. C. KALAVATHEY INSPECTOR 20 Uttar Pradesh Shri INDU BHUSHAN NAUTIYAL SI 21 Uttar Pradesh Shri OM PRAKASH ARYA INSPECTOR 22 West Bengal Shri PARTHA PRATIM GUHA ASI 23 West Bengal Shri PURNA CHANDRA DUTTA ASI PC OPERATION & OFFICE AUTOMATION From 01/05/2000 to 12/05/2000 1 Andhra Pradesh Shri LALSAHEB BANDANAPUDI DY.SP 2 Andhra Pradesh Shri V. RUDRA KUMAR DY.SP 3 Border Security Force Shri ASHOK ARJUN PATIL DY.COMDT. 4 Border Security Force Shri DANIEL ADHIKARI DY.COMDT. 5 Border Security Force Shri DR. VINAYA BHARATI CMO 6 CISF Shri JISHNU PRASANNA MUKHERJEE ASST.COMDT. 7 CISF Shri K.K. SHARMA ASST.COMDT. -

Nominations for Padma Awards 2011
c Nominations fof'P AWARDs 2011 ADMA ~ . - - , ' ",::i Sl. Name';' Field State No ShriIshwarappa,GurapJla Angadi Art Karnataka " Art-'Cinema-Costume Smt. Bhanu Rajopadhye Atharya Maharashtra 2. Designing " Art - Hindustani 3. Dr; (Smt.).Prabha Atre Maharashtra , " Classical Vocal Music 4. Shri Bhikari.Charan Bal Art - Vocal Music 0, nssa·' 5. Shri SamikBandyopadhyay Art - Theatre West Bengal " 6: Ms. Uttara Baokar ',' Art - Theatre , Maharashtra , 7. Smt. UshaBarle Art Chhattisgarh 8. Smt. Dipali Barthakur Art " Assam Shri Jahnu Barua Art - Cinema Assam 9. , ' , 10. Shri Neel PawanBaruah Art Assam Art- Cinema Ii. Ms. Mubarak Begum Rajasthan i", Playback Singing , , , 12. ShriBenoy Krishen Behl Art- Photography Delhi " ,'C 13. Ms. Ritu Beri , Art FashionDesigner Delhi 14. Shri.Madhur Bhandarkar Art - Cinema Maharashtra Art - Classical Dancer IS. Smt. Mangala Bhatt Andhra Pradesh Kathak Art - Classical Dancer 16. ShriRaghav Raj Bhatt Andhra Pradesh Kathak : Art - Indian Folk I 17., Smt. Basanti Bisht Uttarakhand Music Art - Painting and 18. Shri Sobha Brahma Assam Sculpture , Art - Instrumental 19. ShriV.S..K. Chakrapani Delhi, , Music- Violin , PanditDevabrata Chaudhuri alias Debu ' Art - Instrumental 20. , Delhi Chaudhri ,Music - Sitar 21. Ms. Priyanka Chopra Art _Cinema' Maharashtra 22. Ms. Neelam Mansingh Chowdhry Art_ Theatre Chandigarh , ' ,I 23. Shri Jogen Chowdhury Art- Painting \VesfBengal 24.' Smt. Prafulla Dahanukar Art ~ Painting Maharashtra ' . 25. Ms. Yashodhara Dalmia Art - Art History Delhi Art - ChhauDance 26. Shri Makar Dhwaj Darogha Jharkhand Seraikella style 27. Shri Jatin Das Art - Painting Delhi, 28. Shri ManoharDas " Art Chhattisgarh ' 29. , ShriRamesh Deo Art -'Cinema ,Maharashtra Art 'C Hindustani 30. Dr. Ashwini Raja Bhide Deshpande Maharashtra " classical vocalist " , 31. ShriDeva Art - Music Tamil Nadu Art- Manipuri Dance 32. -
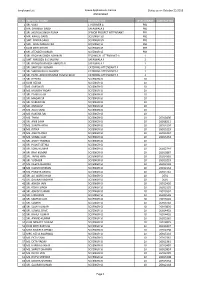
Employee List for Rti As on 25-Oct-2018
Employee List Space Applications Centre Status as on October 25,2018 Ahmedabad Sr.No. EMPLOYEE NAME DESIGNATION LEVEL/GRADE CONTACT NO 1 SRI. VIJAY L V DRIVER A PB1 2 MR. SHANKER SINGH SAFAIWALA B PB1 3 SRI. JAGDISH SINGH PUNIA SENIOR PROJECT ATTENDANT PB1 4 MR. RAHUL GARG SCI/ENGR-SD PB3 5 SMT. BINDIA SAHU SCI/ENGR-SD PB3 6 SMT. ANJU MALHOTRA SCI/ENGR-SE PB3 7 KUM KRITI KHATRI SCI/ENGR-SE PB3 8 SRI. JITENDER KUMAR SCI/ENGR-SE PB3 9 SRI. BACHAN SINGH ADHIKARI TECHNICAL ATTENDANT-A 1 10 SMT. NIRUBEN B CHAUHAN SAFAIWALA A 1 11 SRI. RATHOD MANISH AMRUTLAL SAFAIWALA A 1 12 SRI. SANTOSH KUMAR CATERING ATTENDANT-A 1 13 SRI. SHIRISH BALU GHARDE CATERING ATTENDANT-A 1 14 SRI. PATEL AKSHAYKUMAR YOGESHBHAI CATERING ATTENDANT-A 1 15 SRI. DEEPAK SCI/ENGR-SC 10 16 KUM HEENA SCI/ENGR-SC 10 17 MS. SARSWATI SCI/ENGR-SC 10 18 MS. MONIKA YADAV SCI/ENGR-SC 10 19 SRI. PANKAJ JAIN SCI/ENGR-SC 10 20 SRI. MADAR SK SCI/ENGR-SC 10 21 SRI. VARENYAM SCI/ENGR-SC 10 22 SRI. JISHAD M SCI/ENGR-SC 10 23 MS. ALKA SAINI SCI/ENGR-SC 10 24 MS. RAKSHA RAI SCI/ENGR-SC 10 25 MS. TANVI SCI/ENGR-SC 10 26918406 26 SRI. ANIK SAHA SCI/ENGR-SC 10 26918211 27 MS. SUNITA ARYA SCI/ENGR-SC 10 26914194 28 MS. RITIKA SCI/ENGR-SC 10 26915219 29 MS. ANKITA RANI SCI/ENGR-SC 10 26915234 30 MS. SONALI JAIN SCI/ENGR-SC 10 26915234 31 SRI. -

Solving the Global Cooling Challenge How to Counter the Climate Threat from Room Air Conditioners
Solving the Global Cooling Challenge How to Counter the Climate Threat from Room Air Conditioners By Iain Campbell, Ankit Kalanki, and Sneha Sachar A report by M OUN KY T C A I O N R I N E STIT U T Authors & Acknowledgments Authors Iain Campbell, Ankit Kalanki, and Sneha Sachar (AEEE) * Authors listed alphabetically. All authors from Rocky Mountain Institute unless otherwise noted. Contacts Iain Campbell, [email protected] Ankit Kalanki, [email protected] Suggested Citation Sachar, Sneha, Iain Campbell, and Ankit Kalanki, Solving the Global Cooling Challenge: How to Counter the Climate Threat from Room Air Conditioners. Rocky Mountain Institute, 2018. www.rmi.org/insight/solving_the_global_cooling_challenge. Editorial/Design Editorial Director: Cindie Baker Editor: Laurie Guevara-Stone Layout: Aspire Design, New Delhi Images courtesy of iStock unless otherwise noted. Acknowledgments The authors thank the following individuals/organizations for offering their insights and perspectives on this work. The names are listed in alphabetical order by last name. Omar Abdelaziz, Clean Energy Air and Water Technologies, FZE Paul Bunje, Conservation X Labs Alex Dehgan, Conservation X labs Sukumar Devotta, Former Director at National Environmental Engineering Research Institute Gabrielle Dreyfus, Institute of Governance and Sustainable Development John Dulac, International Energy Agency Chad Gallinat, Conservation X Labs Ben Hartley, SEforALL Saikiran Kasamsetty, Alliance for an Energy Efficient Economy Satish Kumar, Alliance for an Energy Efficient -

Standing Committee on Nutrition
UNITED NATIONS SYSTEM Standing Committee on Nutrition http://www.unsystem.org/scn/ Report of the Standing Committee on Nutrition at its Thirtieth Session Hosted by the M S Swaminathan Research Foundation (MSSRF) at the Indian Institute of Technology Madras Chennai, India, 3-7 March 2003 A. Introduction ........................................................................................................................................... 1 B. Symposium on Mainstreaming Nutrition to Improve Development Outcomes ....................... 2 C. Working Group meetings, summary reports .................................................................................... 2 C.1 Nutrition Throughout the Lifecycle. ......................................................................................... 2 C.2 Nutrition in Emergencies ............................................................................................................ 3 C.3 Nutrition and HIV/AIDS........................................................................................................... 3 C.4 Micronutrients ............................................................................................................................... 4 C.5 Nutrition, Ethics and Human Rights......................................................................................... 4 C.6 Capacity Development in Food and Nutrition........................................................................ 5 C.7 Nutrition of School Age Children.............................................................................................