The Emerging Role of GATA Transcription Factors in Development and Disease
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of PU.1 and GATA-1 Transcription Factors During Normal and Leukemogenic Hematopoiesis
Leukemia (2010) 24, 1249–1257 & 2010 Macmillan Publishers Limited All rights reserved 0887-6924/10 www.nature.com/leu REVIEW The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis P Burda1, P Laslo2 and T Stopka1,3 1Department of Pathophysiology and Center of Experimental Hematology, First Faculty of Medicine, Charles University, Prague, Czech Republic; 2Section of Experimental Haematology, Leeds Institute of Molecular Medicine, University of Leads, St James’s University Hospital, Leeds, UK and 31st Department of Medicine-Hematology, General University Hospital, Prague, Czech Republic Hematopoiesis is coordinated by a complex regulatory network Additional domains include an N-terminal acidic domain and a of transcription factors and among them PU.1 (Spi1, Sfpi1) glutamine-rich domain, both involved in transcriptional activa- represents a key molecule. This review summarizes the tion, as well as a PEST domain involved in protein–protein indispensable requirement of PU.1 during hematopoietic cell fate decisions and how the function of PU.1 can be modulated interactions. PU.1 protein can be modified post-translationally by protein–protein interactions with additional factors. The by phosporylation at serines 41 (N-terminal acidic domain) and mutual negative regulation between PU.1 and GATA-1 is 142 and 148 (PEST domain), which results in augmented detailed within the context of normal and leukemogenic activity. hematopoiesis and the concept of ‘differentiation therapy’ to The PU.1 protein can physically interact with a variety of restore normal cellular differentiation of leukemic cells is regulatory factors including (i) general transcription factors discussed. Leukemia (2010) 24, 1249–1257; doi:10.1038/leu.2010.104; (TFIID, TBP), (ii) early hematopoietic transcription factors published online 3 June 2010 (GATA-2 and Runx-1), (iii) erythroid factor (GATA-1) and (iv) Keywords: PU.1; leukemia differentiation; GATA-1; chromatin; non-erythroid factors (C/EBPa, C/EBPb, IRF4/8 and c-Jun). -

The Ageing Haematopoietic Stem Cell Compartment
REVIEWS The ageing haematopoietic stem cell compartment Hartmut Geiger1,2, Gerald de Haan3 and M. Carolina Florian1 Abstract | Stem cell ageing underlies the ageing of tissues, especially those with a high cellular turnover. There is growing evidence that the ageing of the immune system is initiated at the very top of the haematopoietic hierarchy and that the ageing of haematopoietic stem cells (HSCs) directly contributes to changes in the immune system, referred to as immunosenescence. In this Review, we summarize the phenotypes of ageing HSCs and discuss how the cell-intrinsic and cell-extrinsic mechanisms of HSC ageing might promote immunosenescence. Stem cell ageing has long been considered to be irreversible. However, recent findings indicate that several molecular pathways could be targeted to rejuvenate HSCs and thus to reverse some aspects of immunosenescence. HSC niche The current demographic shift towards an ageing popu- The innate immune system is also affected by ageing. A specialized lation is an unprecedented global phenomenon that has Although an increase in the number of myeloid precur- microenvironment that profound implications. Ageing is associated with tissue sors has been described in the bone marrow of elderly interacts with haematopoietic attrition and an increased incidence of many types of can- people, the oxidative burst and the phagocytic capacity of stem cells (HSCs) to regulate cers, including both myeloid and lymphoid leukaemias, and both macrophages and neutrophils are decreased in these their fate. other haematopoietic cell malignancies1,2. Thus, we need individuals12,13. Moreover, the levels of soluble immune to understand the molecular and cellular mechanisms of mediators are altered with ageing. -

Genomic Analysis of Bone Marrow Failure and Myelodysplastic Syndromes Reveals Phenotypic and Diagnostic Complexity
Bone Marrow Failure SUPPLEMENTARY APPENDIX Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity Michael Y. Zhang,1 Siobán B. Keel,2 Tom Walsh,3 Ming K. Lee,3 Suleyman Gulsuner,3 Amanda C. Watts,3 Colin C. Pritchard,4 Stephen J. Salipante,4 Michael R. Jeng,5 Inga Hofmann,6 David A. Williams,6,7 Mark D. Fleming,8 Janis L. Abkowitz,2 Mary-Claire King,3 and Akiko Shimamura1,9,10 M.Y.Z. and S.B.K. contributed equally to this work. 1Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA; 2Department of Medicine, Division of Hematology, University of Washington, Seattle, WA; 3Department of Medicine and Department of Genome Sciences, University of Washington, Seattle, WA; 4Department of Laboratory Medicine, University of Washington, Seattle, WA; 5Department of Pediatrics, Stanford Uni- versity School of Medicine, Stanford, CA; 6Division of Hematology/Oncology, Boston Children’s Hospital, Dana Farber Cancer Insti- tute, and Harvard Medical School, Boston, MA; 7Harvard Stem Cell Institute, Boston, MA; 8Department of Pathology, Boston Children’s Hospital, MA; 9Department of Pediatric Hematology/Oncology, Seattle Children’s Hospital, WA; 10Department of Pedi- atrics, University of Washington, Seattle, WA, USA ©2014 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2014.113456 Manuscript received on July 22, 2014. Manuscript accepted on September 15, 2014. Correspondence: [email protected] Supplementary Methods Genomics. Libraries were prepared in 96-well format with a Bravo liquid handling robot (Agilent Technologies). One to two micrograms of genomic DNA were sheared to a peak size of 150 bp using a Covaris E series instrument. -

GATA3 As an Adjunct Prognostic Factor in Breast Cancer Patients with Less Aggressive Disease: a Study with a Review of the Literature
diagnostics Article GATA3 as an Adjunct Prognostic Factor in Breast Cancer Patients with Less Aggressive Disease: A Study with a Review of the Literature Patrizia Querzoli 1, Massimo Pedriali 1 , Rosa Rinaldi 2 , Paola Secchiero 3, Paolo Giorgi Rossi 4 and Elisabetta Kuhn 5,6,* 1 Section of Anatomic Pathology, Department of Morphology, Surgery and Experimental Medicine, University of Ferrara, 44124 Ferrara, Italy; [email protected] (P.Q.); [email protected] (M.P.) 2 Section of Anatomic Pathology, ASST Mantova, Ospedale Carlo Poma, 46100 Mantova, Italy; [email protected] 3 Surgery and Experimental Medicine and Interdepartmental Center of Technology of Advanced Therapies (LTTA), Department of Morphology, University of Ferrara, 44121 Ferrara, Italy; [email protected] 4 Epidemiology Unit, Azienda Unità Sanitaria Locale-IRCCS di Reggio Emilia, 42122 Reggio Emilia, Italy; [email protected] 5 Division of Pathology, Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, 20122 Milano, Italy 6 Department of Biomedical, Surgical, and Dental Sciences, University of Milan, 20122 Milano, Italy * Correspondence: [email protected]; Tel.: +39-02-5032-0564; Fax: +39-02-5503-2860 Abstract: Background: GATA binding protein 3 (GATA3) expression is positively correlated with Citation: Querzoli, P.; Pedriali, M.; estrogen receptor (ER) expression, but its prognostic value as an independent factor remains unclear. Rinaldi, R.; Secchiero, P.; Rossi, P.G.; Thus, we undertook the current study to evaluate the expression of GATA3 and its prognostic value Kuhn, E. GATA3 as an Adjunct in a large series of breast carcinomas (BCs) with long-term follow-up. Methods: A total of 702 Prognostic Factor in Breast Cancer consecutive primary invasive BCs resected between 1989 and 1993 in our institution were arranged Patients with Less Aggressive in tissue microarrays, immunostained for ER, progesterone receptor (PR), ki-67, HER2, p53, and Disease: A Study with a Review of GATA3, and scored. -
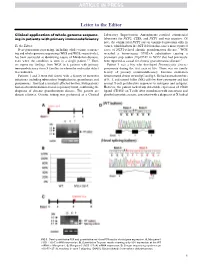
Clinical Application of Whole-Genome Sequencing in Patients with Primary
Letter to the Editor Clinical application of whole-genome sequenc- Laboratory Improvement Amendments–certified commercial ing in patients with primary immunodeficiency laboratory for NCF2, CYBA, and NCF1 and was negative. Of note, the commercial NCF1 screen examined mutations only in To the Editor: exon 2, which harbors the 2GT deletion that causes most reported Next-generation sequencing, including whole-exome sequenc- cases of NCF1-related chronic granulomatous disease.4 WGS ing and whole-genome sequencing (WES and WGS, respectively), revealed a homozygous 579G>A substitution causing a has been successful at identifying causes of Mendelian diseases, premature stop codon (Trp193X) in NCF1 that had previously even when the condition is seen in a single patient.1-3 Here, been reported as causal for chronic granulomatous disease.5 we report our findings from WGS in 6 patients with primary Patient 3 was a boy who developed Pneumocystis jiroveci immunodeficiency from 5 families in whom the molecular defect pneumonia during the first year of life. There was no family was unknown. history of primary immunodeficiency. Immune evaluation Patients 1 and 2 were full sisters with a history of recurrent demonstrated absent serum IgG and IgA. He had normal numbers infections, including tuberculous lymphadenitis, granulomas, and of B, T, and natural killer (NK) cells by flow cytometry and had pneumonias. They had a similarly affected brother. Both patients normal T-cell proliferative responses to mitogens and antigens. had an absent rhodamine-based respiratory burst, confirming the However, the patient lacked any detectable expression of CD40 diagnosis of chronic granulomatous disease. The parents are ligand (CD154) on T cells after stimulation with ionomycin and distant relatives. -

Diverse T-Cell Differentiation Potentials of Human Fetal Thymus, Fetal Liver, Cord Blood and Adult Bone Marrow CD34 Cells On
IMMUNOLOGY ORIGINAL ARTICLE Diverse T-cell differentiation potentials of human fetal thymus, fetal liver, cord blood and adult bone marrow CD34 cells on lentiviral Delta-like-1-modified mouse stromal cells Ekta Patel,1 Bei Wang,1 Lily Lien,2 Summary Yichen Wang,2 Li-Jun Yang,3 Jan Human haematopoietic progenitor/stem cells (HPCs) differentiate into S. Moreb4 and Lung-Ji Chang1 functional T cells in the thymus through a series of checkpoints. A conve- 1Department of Molecular Genetics and nient in vitro system will greatly facilitate the understanding of T-cell Microbiology, College of Medicine, University of Florida, Gainesville, FL, USA, 2Vectorite development and future engineering of therapeutic T cells. In this report, Biomedica Inc., Taipei, Taiwan, 3Department we established a lentiviral vector-engineered stromal cell line (LSC) of Pathology, Immunology and Laboratory expressing the key lymphopoiesis regulator Notch ligand, Delta-like 1 Medicine, University of Florida, Gainesville, (DL1), as feeder cells (LSC-mDL1) supplemented with Flt3 ligand (fms- 4 FL, USA, and Department of Medicine, like tyrosine kinase 3, Flt3L or FL) and interleukin-7 for the development University of Florida, Gainesville, FL, USA of T cells from CD34+ HPCs. We demonstrated T-cell development from human HPCs with various origins including fetal thymus (FT), fetal liver (FL), cord blood (CB) and adult bone marrow (BM). The CD34+ HPCs from FT, FL and adult BM expanded more than 100-fold before reaching the b-selection and CD4/CD8 double-positive T-cell stage. The CB HPCs, on the other hand, expanded more than 1000-fold before b-selection. -

A Core Transcriptional Network Composed of Pax2/8, Gata3 and Lim1 Regulates Key Players of Pro/Mesonephros Morphogenesis
Developmental Biology 382 (2013) 555–566 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/locate/developmentalbiology Genomes and Developmental Control A core transcriptional network composed of Pax2/8, Gata3 and Lim1 regulates key players of pro/mesonephros morphogenesis Sami Kamel Boualia a, Yaned Gaitan a, Mathieu Tremblay a, Richa Sharma a, Julie Cardin b, Artur Kania b, Maxime Bouchard a,n a Goodman Cancer Research Centre and Department of Biochemistry, McGill University, 1160 Pine Ave. W., Montreal, Quebec, Canada H3A 1A3 b Institut de Recherches Cliniques de Montréal, Montréal, Québec, Canada H2W 1R7, Department of Anatomy and Cell Biology, Division of Experimental Medicine, McGill University, Montréal, Quebec, Canada, H3A 2B2 and Faculté de médecine, Université de Montréal, Montréal, Quebec, Canada, H3C 3J7. article info abstract Article history: Translating the developmental program encoded in the genome into cellular and morphogenetic Received 23 January 2013 functions requires the deployment of elaborate gene regulatory networks (GRNs). GRNs are especially Received in revised form crucial at the onset of organ development where a few regulatory signals establish the different 27 July 2013 programs required for tissue organization. In the renal system primordium (the pro/mesonephros), Accepted 30 July 2013 important regulators have been identified but their hierarchical and regulatory organization is still Available online 3 August 2013 elusive. Here, we have performed a detailed analysis of the GRN underlying mouse pro/mesonephros Keywords: development. We find that a core regulatory subcircuit composed of Pax2/8, Gata3 and Lim1 turns on a Kidney development deeper layer of transcriptional regulators while activating effector genes responsible for cell signaling Transcription and tissue organization. -

The Title of the Dissertation
UNIVERSITY OF CALIFORNIA SAN DIEGO Novel network-based integrated analyses of multi-omics data reveal new insights into CD8+ T cell differentiation and mouse embryogenesis A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Bioinformatics and Systems Biology by Kai Zhang Committee in charge: Professor Wei Wang, Chair Professor Pavel Arkadjevich Pevzner, Co-Chair Professor Vineet Bafna Professor Cornelis Murre Professor Bing Ren 2018 Copyright Kai Zhang, 2018 All rights reserved. The dissertation of Kai Zhang is approved, and it is accept- able in quality and form for publication on microfilm and electronically: Co-Chair Chair University of California San Diego 2018 iii EPIGRAPH The only true wisdom is in knowing you know nothing. —Socrates iv TABLE OF CONTENTS Signature Page ....................................... iii Epigraph ........................................... iv Table of Contents ...................................... v List of Figures ........................................ viii List of Tables ........................................ ix Acknowledgements ..................................... x Vita ............................................. xi Abstract of the Dissertation ................................. xii Chapter 1 General introduction ............................ 1 1.1 The applications of graph theory in bioinformatics ......... 1 1.2 Leveraging graphs to conduct integrated analyses .......... 4 1.3 References .............................. 6 Chapter 2 Systematic -

5045.Full.Pdf
IFN Consensus Sequence Binding Protein (Icsbp) Is Critical for Eosinophil Development This information is current as Maja Milanovic, Grzegorz Terszowski, Daniela Struck, of September 28, 2021. Oliver Liesenfeld and Dirk Carstanjen J Immunol 2008; 181:5045-5053; ; doi: 10.4049/jimmunol.181.7.5045 http://www.jimmunol.org/content/181/7/5045 Downloaded from References This article cites 47 articles, 33 of which you can access for free at: http://www.jimmunol.org/content/181/7/5045.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 28, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2008 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology IFN Consensus Sequence Binding Protein (Icsbp) Is Critical for Eosinophil Development1 Maja Milanovic,2* Grzegorz Terszowski,2† Daniela Struck,2‡ Oliver Liesenfeld,‡ and Dirk Carstanjen3* IFN consensus sequence binding protein (Icsbp) (IFN response factor-8) is a hematopoietic transcription factor with dual functions in myelopoiesis and immunity. -

Transcriptome Analysis of Gravitational Effects on Mouse Skeletal Muscles Under Microgravity and Artificial 1 G Onboard Environm
www.nature.com/scientificreports OPEN Transcriptome analysis of gravitational efects on mouse skeletal muscles under microgravity and artifcial 1 g onboard environment Risa Okada1,2, Shin‑ichiro Fujita3,4, Riku Suzuki5,6, Takuto Hayashi3,5, Hirona Tsubouchi5, Chihiro Kato5,7, Shunya Sadaki5, Maho Kanai5,6, Sayaka Fuseya3,5, Yuri Inoue3,5, Hyojung Jeon5, Michito Hamada5, Akihiro Kuno5,6, Akiko Ishii8, Akira Tamaoka8, Jun Tanihata9, Naoki Ito10, Dai Shiba1,2, Masaki Shirakawa1,2, Masafumi Muratani1,4, Takashi Kudo1,5* & Satoru Takahashi1,5* Spacefight causes a decrease in skeletal muscle mass and strength. We set two murine experimental groups in orbit for 35 days aboard the International Space Station, under artifcial earth‑gravity (artifcial 1 g; AG) and microgravity (μg; MG), to investigate whether artifcial 1 g exposure prevents muscle atrophy at the molecular level. Our main fndings indicated that AG onboard environment prevented changes under microgravity in soleus muscle not only in muscle mass and fber type composition but also in the alteration of gene expression profles. In particular, transcriptome analysis suggested that AG condition could prevent the alterations of some atrophy‑related genes. We further screened novel candidate genes to reveal the muscle atrophy mechanism from these gene expression profles. We suggest the potential role of Cacng1 in the atrophy of myotubes using in vitro and in vivo gene transductions. This critical project may accelerate the elucidation of muscle atrophy mechanisms. Gravity is the most constant factor afecting the entire process of evolution of organisms on Earth. As adapting to a changing environment is key for any organism’s survival, the constant mechanical stimulus of gravitational force has been shared by all organisms on Earth through evolution 1. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

GATA2 Regulates the Erythropoietin Receptor in T(12;21) ALL
GATA2 regulates the erythropoietin receptor in t(12;21) ALL Gaine, M. E., Sharpe, D. J., Smith, J. S., Colyer, H., Hodges, V. M., Lappin, T. R., & Mills, K. I. (2017). GATA2 regulates the erythropoietin receptor in t(12;21) ALL. Oncotarget, 8(39), 66061-66074. https://doi.org/10.18632/oncotarget.19792 Published in: Oncotarget Document Version: Publisher's PDF, also known as Version of record Queen's University Belfast - Research Portal: Link to publication record in Queen's University Belfast Research Portal Publisher rights © 2017 The Authors. This is an open access article published under a Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution and reproduction in any medium, provided the author and source are cited. General rights Copyright for the publications made accessible via the Queen's University Belfast Research Portal is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The Research Portal is Queen's institutional repository that provides access to Queen's research output. Every effort has been made to ensure that content in the Research Portal does not infringe any person's rights, or applicable UK laws. If you discover content in the Research Portal that you believe breaches copyright or violates any law, please contact [email protected]. Download date:04. Oct. 2021 www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 39), pp: 66061-66074 Research Paper GATA2 regulates the erythropoietin receptor in t(12;21) ALL Marie E.