Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Inhibition of Breast and Brain Cancer Cell Growth by Bccipα, An
Oncogene (2001) 20, 336 ± 345 ã 2001 Nature Publishing Group All rights reserved 0950 ± 9232/01 $15.00 www.nature.com/onc Inhibition of breast and brain cancer cell growth by BCCIPa,an evolutionarily conserved nuclear protein that interacts with BRCA2 Jingmei Liu1, Yuan Yuan1,2, Juan Huan2 and Zhiyuan Shen*,1 1Department of Molecular Genetics and Microbiology, University of New Mexico Health Sciences Center; 915 Camino de Salud, NE. Albuquerque, New Mexico, NM 87131, USA; 2Graduate Program of Molecular Genetics, College of Medicine, University of Illinois at Chicago, 900 S. Ashland Ave. Chicago, Illinois, IL 60607, USA BRCA2 is a tumor suppressor gene involved in mammary mouse BRCA2. It is expected that important functions tumorigenesis. Although important functions have been of BRCA2 reside in these conserved domains. Based on assigned to a few conserved domains of BRCA2, little is the functional analysis of the conserved BRCA2 known about the longest internal conserved domain domains, several models have been proposed for the encoded by exons 14 ± 24. We identi®ed a novel protein, role of BRCA2 in tumor suppression. designated BCCIPa, that interacts with part of the An N-terminus conserved domain in exon 3 (amino internal conserved region of human BRCA2. Human acids 48 ± 105) has been implicated in transcriptional BCCIP represents a family of proteins that are regulation of gene expression (Milner et al., 1997; evolutionarily conserved, and contain three distinct Nordling et al., 1998). Deletion of this region has been domains: an N-terminus acidic domain (NAD) of 30 ± identi®ed in breast cancers (Nordling et al., 1998). -

TBXA2R Rsnps, Transcriptional Factor Binding Sites and Asthma in Asians
Open Journal of Pediatrics, 2014, 4, 148-161 Published Online June 2014 in SciRes. http://www.scirp.org/journal/ojped http://dx.doi.org/10.4236/ojped.2014.42021 TBXA2R rSNPs, Transcriptional Factor Binding Sites and Asthma in Asians Norman E. Buroker Department of Pediatrics, University of Washington, Seattle, USA Email: [email protected] Received 25 January 2014; revised 20 February 2014; accepted 27 February 2014 Copyright © 2014 by author and Scientific Research Publishing Inc. This work is licensed under the Creative Commons Attribution International License (CC BY). http://creativecommons.org/licenses/by/4.0/ Abstract Four regulatory single nucleotide polymorphisms (rSNPs) (rs2238631, rs2238632, rs2238633 and rs2238634) in intron one, two rSNPs (rs1131882 and rs4523) in exon 3 and one rSNP (rs5756) in the 3’UTR of the thromboxane A2 receptor (TBXA2R) gene have been associated with childhood- onset asthma in Asians. These rSNP alleles alter the DNA landscape for potential transcriptional factors (TFs) to attach resulting in changes in transcriptional factor binding sites (TFBS). These TFBS changes are examined with respect to asthma which has been found to be significantly asso- ciated with the rSNPs. Keywords TBXA2R, rSNPs, TFBS, Asthma 1. Introduction Asthma is a chronic inflammatory condition of the airways characterized by recurrent episodes of reversible air- way obstruction and increased bronchial hyper-responsiveness which results from the interactions between gen- es and environmental factors [1]-[3]. Asthma causes episodes of wheeze, cough, and shortness of breath [4]. Re- cent studies indicate that the genetic factors of childhood-onset asthma differ from those of adult-onset asthma [3] [5]. -

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

A Reciprocal Effort Towards Better Approaches for Drug Discovery
Acta Pharmacologica Sinica (2013) 34: 765–776 npg © 2013 CPS and SIMM All rights reserved 1671-4083/13 $32.00 www.nature.com/aps Review iPSCs and small molecules: a reciprocal effort towards better approaches for drug discovery Ru ZHANG1, Li-hong ZHANG2, Xin XIE1, 2, * 1Laboratory of Receptor-based Bio-medicine, Shanghai Key Laboratory of Signaling and Disease Research, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China; 2CAS Key Laboratory of Receptor Research, the National Center for Drug Screening, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China The revolutionary induced pluripotent stem cell (iPSC) technology provides a new path for cell replacement therapies and drug screen- ing. Patient-specific iPSCs and subsequent differentiated cells manifesting disease phenotypes will finally position human disease pathology at the core of drug discovery. Cells used to test the toxic effects of drugs can also be generated from normal iPSCs and pro- vide a much more accurate and cost-effective system than many animal models. Here, we highlight the recent progress in iPSC-based cell therapy, disease modeling and drug evaluations. In addition, we discuss the use of small molecule drugs to improve the genera- tion of iPSCs and understand the reprogramming mechanism. It is foreseeable that the interplay between iPSC technology and small molecule compounds will push forward the applications of iPSC-based therapy and screening systems in the real world and eventually revolutionize the methods used to treat diseases. Keywords: induced pluripotent stem cells (iPSCs); disease modeling; drug screening; toxicity evaluation; cell replacement therapy; small molecules; drug development Acta Pharmacologica Sinica (2013) 34: 765–776; doi: 10.1038/aps.2013.21; published online 22 Apr 2013 Introduction his/her own iPSCs[1–3]. -

Analysis of Trans Esnps Infers Regulatory Network Architecture
Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Anat Kreimer All rights reserved ABSTRACT Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer eSNPs are genetic variants associated with transcript expression levels. The characteristics of such variants highlight their importance and present a unique opportunity for studying gene regulation. eSNPs affect most genes and their cell type specificity can shed light on different processes that are activated in each cell. They can identify functional variants by connecting SNPs that are implicated in disease to a molecular mechanism. Examining eSNPs that are associated with distal genes can provide insights regarding the inference of regulatory networks but also presents challenges due to the high statistical burden of multiple testing. Such association studies allow: simultaneous investigation of many gene expression phenotypes without assuming any prior knowledge and identification of unknown regulators of gene expression while uncovering directionality. This thesis will focus on such distal eSNPs to map regulatory interactions between different loci and expose the architecture of the regulatory network defined by such interactions. We develop novel computational approaches and apply them to genetics-genomics data in human. We go beyond pairwise interactions to define network motifs, including regulatory modules and bi-fan structures, showing them to be prevalent in real data and exposing distinct attributes of such arrangements. We project eSNP associations onto a protein-protein interaction network to expose topological properties of eSNPs and their targets and highlight different modes of distal regulation. -

Defective Lymphocyte Chemotaxis in Я-Arrestin2- and GRK6-Deficient Mice
Defective lymphocyte chemotaxis in -arrestin2- and GRK6-deficient mice Alan M. Fong*, Richard T. Premont*, Ricardo M. Richardson*, Yen-Rei A. Yu†, Robert J. Lefkowitz*‡§, and Dhavalkumar D. Patel*†¶ Departments of *Medicine, ‡Biochemistry, and †Immunology, and §Howard Hughes Medical Institute, Duke University Medical Center, Durham, NC 27710 Contributed by Robert J. Lefkowitz, April 4, 2002 Lymphocyte chemotaxis is a complex process by which cells move kinase, extracellular receptor kinase, and c-jun terminal kinase within tissues and across barriers such as vascular endothelium and activation (9–12), they might also act as positive regulators of is usually stimulated by chemokines such as stromal cell-derived chemotaxis. To evaluate the role of the GRK-arrestin pathway factor-1 (CXCL12) acting via G protein-coupled receptors. Because in chemotaxis, we studied the chemotactic responses of lym- members of this receptor family are regulated (‘‘desensitized’’) by phocytes from -arrestin- and GRK-deficient mice toward G protein-coupled receptor kinase (GRK)-mediated receptor phos- gradients of stromal cell-derived factor 1 (CXCL12), a well  phorylation and -arrestin binding, we examined signaling and characterized chemokine whose receptor is CXCR4, a core- chemotactic responses in splenocytes derived from knockout mice ceptor for HIV. deficient in various -arrestins and GRKs, with the expectation that these responses might be enhanced. Knockouts of -arrestin2, Materials and Methods GRK5, and GRK6 were examined because all three proteins are :expressed at high levels in purified mouse CD3؉ T and B220؉ B Mice. The following mouse strains were used in this study splenocytes. CXCL12 stimulation of membrane GTPase activity was -arrestin2-deficient (back-crossed for six generations onto the unaffected in splenocytes derived from GRK5-deficient mice but C57͞BL6 background; ref. -

Genotyping for Response to Physical Training
Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2019 Genotyping for Response to Physical Training Stacy Simmons Wright State University Follow this and additional works at: https://corescholar.libraries.wright.edu/etd_all Part of the Molecular Biology Commons Repository Citation Simmons, Stacy, "Genotyping for Response to Physical Training" (2019). Browse all Theses and Dissertations. 2109. https://corescholar.libraries.wright.edu/etd_all/2109 This Thesis is brought to you for free and open access by the Theses and Dissertations at CORE Scholar. It has been accepted for inclusion in Browse all Theses and Dissertations by an authorized administrator of CORE Scholar. For more information, please contact [email protected]. GENOTYPING FOR RESPONSE TO PHYSICAL TRAINING A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By STACY SIMMONS B.S., Wright State University, 2014 _________________________________________________________ 2019 Wright State University WRIGHT STATE UNIVERSITY GRADUATE SCHOOL July 29, 2019 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY Stacy Simmons ENTITLED Genotyping for Response to Physical Training BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science. ___________________________________ Michael Markey, Ph.D. Thesis Director ____________________________________ Madhavi P. Kadakia, Ph.D. Committee on Chair, Department of Biochemistry Final Examination and -

Combined Integrin Phosphoproteomic Analyses and Small Interfering RNA–Based Functional Screening Identify Key Regulators for Cancer Cell Adhesion and Migration
Published OnlineFirst April 7, 2009; DOI: 10.1158/0008-5472.CAN-08-2515 Published Online First on April 7, 2009 as 10.1158/0008-5472.CAN-08-2515 Research Article Combined Integrin Phosphoproteomic Analyses and Small Interfering RNA–Based Functional Screening Identify Key Regulators for Cancer Cell Adhesion and Migration Yanling Chen,1 Bingwen Lu,2 Qingkai Yang,1 Colleen Fearns,3 John R. Yates III,2 and Jiing-Dwan Lee1 Departments of 1Immunology, 2Chemical Physiology, and 3Chemistry, The Scripps Research Institute, La Jolla, California Abstract cytoskeletal components (2, 7) and regulation of cell survival, Integrins interact with extracellular matrix (ECM) and deliver proliferation, differentiation, cell cycle, and migration (8, 9). Understanding the mechanism by which integrins modulate these intracellular signaling for cell proliferation, survival, and motility. During tumor metastasis, integrin-mediated cell cellular activities is of significant biological importance. adhesion to and migration on the ECM proteins are required The most common cancers in human include breast cancer, for cancer cell survival and adaptation to the new microen- prostate cancer, lung cancer, colon cancer, and ovarian cancer (10, 11), and their metastasis is the leading cause of mortality in vironment. Using stable isotope labeling by amino acids in cell cancer patients, causing 90% of deaths from solid tumors (11, 12). culture–mass spectrometry, we profiled the phosphoproteo- During the process of metastasis, tumor cells leave the primary mic changes induced by the interactions of cell integrins with site, travel via blood and/or lymphatic circulatory systems, attach type I collagen, the most common ECM substratum. Integrin- to the substratum of ECM at a distant site, and establish a ECM interactions modulate phosphorylation of 517 serine, secondary tumor, accompanied by angiogenesis of the newly threonine, or tyrosine residues in 513 peptides, corresponding formed neoplasm (12). -

Katalog 2015 Cover Paul Lin *Hinweis Förderung.Indd
Product List 2015 WE LIVE SERVICE Certificates quartett owns two productions sites that are certified according to EN ISO 9001:2008 Quality management systems - Requirements EN ISO 13485:2012 + AC:2012 Medical devices - Quality management systems - Requirements for regulatory purposes GMP Conformity Our quality management guarantees products of highest quality! 2 Foreword to the quartett product list 2015 quartett Immunodiagnostika, Biotechnologie + Kosmetik Vertriebs GmbH welcomes you as one of our new business partners as well as all of our previous loyal clients. You are now member of quartett´s worldwide customers. First of all we would like to introduce ourselves to you. Founded as a family-run company in 1986, quartett ensures for more than a quarter of a century consistent quality of products. Service and support of our valued customers are our daily businesses. And we will continue! In the end 80´s quartett offered radioimmunoassay and enzyme immunoassay kits from different manufacturers in the USA. In the beginning 90´s the company changed its strategy from offering products for routine diagnostic to the increasing field of research and development. Setting up a production plant in 1997 and a second one in 2011 supported this decision. The company specialized its product profile in the field of manufacturing synthetic peptides for antibody production, peptides such as protease inhibitors, biochemical reagents and products for histology, cytology and immunohistology. All products are exclusively manufactured in Germany without outsourcing any production step. Nowadays, we expand into all other diagnostic and research fields and supply our customers in universities, government institutes, pharmaceutical and biotechnological companies, hospitals, and private doctor offices. -

1 Tumor Suppressor PLK2 May Serve As a Biomarker in Triple-Negative Breast Cancer for Improved Response to PLK1 Therapeutics
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448722; this version posted June 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Tumor suppressor PLK2 may serve as a biomarker in triple-negative breast cancer for improved response to PLK1 therapeutics Yang Gao1, 2, 3, Elena B. Kabotyanski1, 2, Elizabeth Villegas7, Jonathan H. Shepherd8, Deanna Acosta1, 2, Clark Hamor1, 2, Tingting Sun2,4,5, Celina Montmeyor-Garcia9, Xiaping He8, Lacey E. Dobrolecki1, 2, 3, Thomas F. Westbrook2, 4, 5, Michael T. Lewis1, 2, 3, Susan G. Hilsenbeck2, 3, Xiang H.-F. Zhang1, 2, 3, 6, Charles M. Perou8 and Jeffrey M. Rosen1, 2 1Department of Molecular and Cellular Biology 2Dan L. Duncan Cancer Center 3Lester and Sue Smith Breast Center 4Department of Molecular and Human Genetics 5Verna & Marrs McLean Department of Biochemistry and Molecular Biology 6McNair Medical Institute Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA 7University of Houston-Downtown, Houston, TX 77002, USA 8The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA 9 Canadian Blood Services, Toronto, ON M5G 2M1, Canada Correspondence to Jeffrey M. Rosen (Mail Stop: BCM130, Room: BCM-M638a, Baylor College of Medicine, 1 Baylor Plaza, Houston, TX 77030. Office: 713-798-6210. Fax: 713-898-8012. Email: [email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448722; this version posted June 16, 2021. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
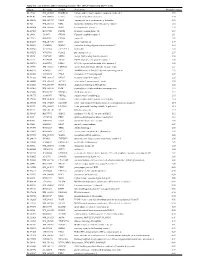
Supplementary Table 1
Table S1. List of Genes Differentially Expressed in YB-1 siRNA-Transfected MCF-7 Cells Unigene Accession Symbol Description Mean fold change Hs.17466 NM_004585 RARRES3 retinoic acid receptor responder (tazarotene induced) 3 3.57 Hs.64746 NM_004669 CLIC3 chloride intracellular channel 3 3.33 Hs.516155 NM_001747 CAPG capping protein (actin filament), gelsolin-like 2.88 Hs.926 NM_002463 MX2 myxovirus (influenza virus) resistance 2 (mouse) 2.72 Hs.643494 NM_005410 SEPP1 selenoprotein P, plasma, 1 2.70 Hs.267038 BC017500 POF1B premature ovarian failure 1B 2.67 Hs.20961 U63917 GPR30 G protein- coupled receptor 30 2532.53 Hs.199877 BE645967 CPNE4 copine IV 2.49 Hs.632177 NM_017458 MVP major vault protein 2.48 Hs.528836 AA005023 NOD27 nucleotide-binding oligomerization domains 27 2.43 Hs.360940 AL589866 dJ222E13.1 kraken-like 2.40 Hs.515575 AI743780 PLAC2 placenta-specific 2 2.37 Hs.25674 AI827820 MBD2 methyl-CpG binding domain protein 2 2.36 Hs.12229 AA149594 TIEG2 TGFB inducible early growth response 2 2.33 Hs.482730 AA053711 EDIL3 EGF-like repeats and discoidin I-like domains 3 2.32 Hs. 370771 NM_000389 CDKN1A cyclin- depen dent kinase in hibitor 1A (p 21, Cip 1) 2282.28 Hs.469175 AI341537 JFC1 NADPH oxidase-related, C2 domain-containing protein 2.28 Hs.514821 AF043341 CCL5 chemokine (C-C motif) ligand 5 2.27 Hs.591292 NM_023915 GPR87 G protein-coupled receptor 87 2.26 Hs.500483 NM_001613 ACTA2 actin, alpha 2, smooth muscle, aorta 2.24 Hs.632824 NM_006729 DIAPH2 diaphanous homolog 2 (Drosophila) 2.22 Hs.369430 NM_000919 PAM peptidylglycine