Viral Respiratory Infections in Adults Focus on RSV and Rapid Molecular Diagnostic Testing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Optumrx Brand Pipeline Forecast
RxOutlook® 1st Quarter 2019 OptumRx brand pipeline forecast Route of Regulatory Estimated Specialty Orphan Drug name Generic name Company Drug class Therapeutic use administration status release date drug drug 2019 Possible launch date Ophthalmological DS-300 DS-300 Eton undisclosed SC Filed NDA 2019 unknown N disease anti-sclerostin Evenity romosozumab Amgen Osteoporosis SC Filed NDA 2/2019 Y N monoclonal antibody tetrahydrofolate iclaprim iclaprim Motif Bio Bacterial infections IV Filed NDA 2/13/2019 Y Y dehydrogenase inhibitor tazarotene/ IDP-118 Valeant retinoid/ corticosteroid Psoriasis TOP Filed NDA 2/15/2019 N N halobetasol adenosine deaminase Mavenclad cladribine Merck/ Teva resistant Multiple sclerosis PO Filed NDA 2/15/2019 Y N deoxyadenosine analog Lotemax Gel loteprednol Valeant corticosteroid Ocular inflammation OP Filed NDA 2/25/2019 N N Nex Gen etabonate turoctocog alfa glyco-PEGylated factor NN-7088 Novo Nordisk Hemophilia IV/SC Filed BLA 2/27/2019 Y N pegol VIII derivative selective sphingosine-1 BAF-312 siponimod Novartis phosphate receptor Multiple sclerosis PO Filed NDA 3/1/2019 Y N agonist midazolam midazolam UCB benzodiazepine Seizures Intranasal Filed NDA 3/1/2019 N Y (USL-261) XeriSol glucagon Xeris glucagon analog Diabetes mellitus SC Filed NDA 3/1/2019 N N Glucagon optum.com/optumrx 1 RxOutlook® 1st Quarter 2019 Route of Regulatory Estimated Specialty Orphan Drug name Generic name Company Drug class Therapeutic use administration status release date drug drug dopamine receptor JZP-507 sodium oxybate Jazz Narcolepsy -

Microorganisms-07-00521-V2.Pdf
microorganisms Review Are Community Acquired Respiratory Viral Infections an Underestimated Burden in Hematology Patients? Cristian-Marian Popescu 1,* , Aurora Livia Ursache 2, Gavriela Feketea 1 , Corina Bocsan 3, Laura Jimbu 1, Oana Mesaros 1, Michael Edwards 4,5, Hongwei Wang 6, Iulia Berceanu 7, Alexandra Neaga 1 and Mihnea Zdrenghea 1,7,* 1 Department of Hematology, Iuliu Hatieganu University of Medicine and Pharmacy, 8 Babes str., 400012 Cluj-Napoca, Romania; [email protected] (G.F.); [email protected] (L.J.); [email protected] (O.M.); [email protected] (A.N.) 2 The Faculty of Veterinary Medicine, University of Agricultural Sciences and Veterinary Medicine, Calea Mănăs, tur 3-5, 400372 Cluj-Napoca, Romania; [email protected] 3 Department of Pharmacology, Toxicology and Clinical Pharmacology, Iuliu Ha¸tieganuUniversity of Medicine and Pharmacy, 400337 Cluj-Napoca, Romania; [email protected] 4 National Heart and Lung Institute, St Mary’s Campus, Imperial College London, London W2 1PG, UK; [email protected] 5 Medical Research Council (MRC) and Asthma UK Centre in Allergic Mechanisms of Asthma, London W2 1PG, UK 6 School of Medicine, Nanjing University, 22 Hankou Road, Nanjing 210093, China; [email protected] 7 Department of Hematology, Ion Chiricuta Oncology Institute, 34-36 Republicii Street, 400015 Cluj-Napoca, Romania; [email protected] * Correspondence: [email protected] (C.-M.P.); [email protected] (M.Z.) Received: 8 October 2019; Accepted: 31 October 2019; Published: 2 November 2019 Abstract: Despite a plethora of studies demonstrating significant morbidity and mortality due to community-acquired respiratory viral (CRV) infections in intensively treated hematology patients, and despite the availability of evidence-based guidelines for the diagnosis and management of respiratory viral infections in this setting, there is no uniform inclusion of respiratory viral infection management in the clinical hematology routine. -
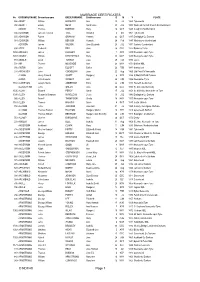
MARRIAGE CERTIFICATES © NDFHS Page 1
MARRIAGE CERTIFICATES No GROOMSURNAME Groomforename BRIDESURNAME Brideforename D M Y PLACE 588 ABBOT William HADAWAY Ann 25 Jul 1869 Tynemouth 935 ABBOTT Edwin NESS Sarah Jane 20 JUL 1882 Wallsend Parrish Church Northumbrland ADAMS Thomas BORTON Mary 16 OCT 1849 Coughton Northampton 556 ADAMSON James Frederick TATE Annabell 6 Oct 1861 Tynemouth 655 ADAMSON Robert GRAHAM Hannah 23 OCT 1847 Darlington Co Durham 581 ADAMSON William BENSON Hannah 24 Feb 1847 Whitehaven Cumberland ADDISON James WILSON Jane Elizabeth 23 JUL 1871 Carlisle, Cumberland 694 ADDY Frederick BELL Jane 26 DEC 1922 Barnsley Yorks 1456 AFFLECK James LUCKLEY Ann 1 APR 1839 Newcastle upon Tyne 1457 AGNEW William KIRKPATRICK Mary 30 MAY 1887 Newcastle upon Tyne 751 AINGER David TURNER Eliza 28 FEB 1870 Essex 704 AIR Thomas MCKENZIE Ann 24 MAY 1871 Belford NBL 936 AISTON John ELLIOTT Esther 26 FEB 1881 Sunderland 244 AITCHISON John COCKBURN Jane 22 Aug 1865 Utd Pres Ch Newcastle ALBION Henry Edward SCOTT Margaret 6 APR 1884 St Mark Millfield Durham ALDER John Cowens WRIGHT Ann 24 JUN 1856 Newcastle /Tyne 1160 ALDERSON Joseph Henry ANDERSON Eliza 22 JUN 1897 Heworth Co Durham ALLABURTON John GREEN Jane 24 DEC 1842 St. Giles ,Durham City 1505 ALLAN Edward PERCY Sarah 17 JUL 1854 St. Nicholas, Newcastle on Tyne 1390 ALLEN Alexander Bowman WANDLESS Jessie 10 JUL 1943 Darlington Co Durham 992 ALLEN Peter F THOMPSON Sheila 18 MAY 1957 Newcastle upon Tyne 1161 ALLEN Thomas HIGGINS Annie 4 OCT 1887 South Shields 158 ALLISON John JACKSON Jane Ann 31 Jul 1859 Colliery, Catchgate, -

Northumberland and Durham Family History Society Unwanted
Northumberland and Durham Family History Society baptism birth marriage No Gsurname Gforename Bsurname Bforename dayMonth year place death No Bsurname Bforename Gsurname Gforename dayMonth year place all No surname forename dayMonth year place Marriage 933ABBOT Mary ROBINSON James 18Oct1851 Windermere Westmorland Marriage 588ABBOT William HADAWAY Ann 25 Jul1869 Tynemouth Marriage 935ABBOTT Edwin NESS Sarah Jane 20 Jul1882 Wallsend Parrish Church Northumbrland Marriage1561ABBS Maria FORDER James 21May1861 Brooke, Norfolk Marriage 1442 ABELL Thirza GUTTERIDGE Amos 3 Aug 1874 Eston Yorks Death 229 ADAM Ellen 9 Feb 1967 Newcastle upon Tyne Death 406 ADAMS Matilda 11 Oct 1931 Lanchester Co Durham Marriage 2326ADAMS Sarah Elizabeth SOMERSET Ernest Edward 26 Dec 1901 Heaton, Newcastle upon Tyne Marriage1768ADAMS Thomas BORTON Mary 16Oct1849 Coughton Northampton Death 1556 ADAMS Thomas 15 Jan 1908 Brackley, Norhants,Oxford Bucks Birth 3605 ADAMS Sarah Elizabeth 18 May 1876 Stockton Co Durham Marriage 568 ADAMSON Annabell HADAWAY Thomas William 30 Sep 1885 Tynemouth Death 1999 ADAMSON Bryan 13 Aug 1972 Newcastle upon Tyne Birth 835 ADAMSON Constance 18 Oct 1850 Tynemouth Birth 3289ADAMSON Emma Jane 19Jun 1867Hamsterley Co Durham Marriage 556 ADAMSON James Frederick TATE Annabell 6 Oct 1861 Tynemouth Marriage1292ADAMSON Jane HARTBURN John 2Sep1839 Stockton & Sedgefield Co Durham Birth 3654 ADAMSON Julie Kristina 16 Dec 1971 Tynemouth, Northumberland Marriage 2357ADAMSON June PORTER William Sidney 1May 1980 North Tyneside East Death 747 ADAMSON -

Repurposing Nucleoside Analogs for Human Coronaviruses
ANTIVIRAL AGENTS crossm Repurposing Nucleoside Analogs for Human Coronaviruses Keivan Zandi,a,b Franck Amblard,a,b Katie Musall,a,b Jessica Downs-Bowen,a,b Ruby Kleinbard,a,b Adrian Oo,a,b Dongdong Cao,c Bo Liang,c Olivia O. Russell,a,b Tamara McBrayer,a,b Leda Bassit,a,b Baek Kim,a,b Raymond F. Schinazia,b aCenter for AIDS Research, Laboratory of Biochemical Pharmacology, Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA Downloaded from bChildren’s Healthcare of Atlanta, Atlanta, Georgia, USA cDepartment of Biochemistry, Emory University School of Medicine, Atlanta, Georgia, USA ABSTRACT Coronavirus disease 2019 (COVID-19) is a serious illness caused by se- vere acute respiratory syndrome coronavirus 2 (SARS-CoV-2 or CoV-2). Some reports claimed certain nucleoside analogs to be active against CoV-2 and thus needed con- firmation. Here, we evaluated a panel of compounds and identified novel nucleoside analogs with antiviral activity against CoV-2 and HCoV-OC43 while ruling out others. http://aac.asm.org/ Of significance, sofosbuvir demonstrated no antiviral effect against CoV-2, and its triphosphate did not inhibit CoV-2 RNA polymerase. KEYWORDS SARS-CoV-2, HCoV-OC43, antiviral agents, nucleoside analogs, remdesivir, sofosbuvir, emtricitabine, lamivudine, COVID-19, Coronaviridae oronavirus disease 2019 (COVID-19) has emerged as a global pandemic with Csignificant morbidity and mortality caused by the severe acute respiratory syn- on December 16, 2020 by guest drome coronavirus 2 (SARS-CoV-2 or CoV-2). CoV-2-infected individuals usually develop mild to severe flulike symptoms, whereas other individuals (particularly the elderly harboring underlying chronic health complications, such as diabetes and heart disease, or immunocompromised individuals) are particularly prone to develop severe to fatal clinical outcomes (1, 2). -

Respiratory Syncytial Virus
RSV Respiratory syncytial virus RSV (Respiratory syncytial virus) is a leading cause of acute respiratory infections. RSV can exploit host immunity and cause a strong inflammatory response that leads to lung damage and virus dissemination. There is a single RSV serotype with two major antigenic subgroups, A and B. RSV is a non-segmented negative-sense single-stranded enveloped RNA virus that belongs to the family of Paramyxoviridae, genus Pneumovirus, subfamily Pneumovirinae. Its 10 genes encode 11 proteins since two overlapping open reading frames in the M2 mRNA yield two distinct matrix proteins, M2-1 and M2-2. The viral envelope contains three proteins, the G glycoprotein, the fusion (F) glycoprotein, and the small hydrophobic (SH) protein. The RSV virus comprises five other structural proteins, the large (L) protein, nucleocapsid (N), phosphoprotein (P), matrix (M), and M2-1, and two non-structural proteins (NS1 and NS2). www.MedChemExpress.com 1 RSV Inhibitors Ac-CoA Synthase Inhibitor1 ALS-8112 Cat. No.: HY-104032 Cat. No.: HY-12983 Ac-CoA Synthase Inhibitor1 is a potent, reversible ALS-8112 is a potent and selective respiratory acetate-dependent acetyl-CoA synthetase 2 syncytial virus (RSV) polymerase inhibitor. The (ACSS2) inhibitor with an IC50 of 0.6 µM. Ac-CoA 5'-triphosphate form of ALS-8112 inhibits RSV Synthase Inhibitor1 inhibits the respiratory polymerase with an IC50 of 0.02 μM. syncytial virus (RSV). Purity: 99.23% Purity: 99.97% Clinical Data: No Development Reported Clinical Data: No Development Reported Size: 10 mM × 1 mL, 5 mg, 10 mg, 25 mg, 50 mg Size: 10 mM × 1 mL, 1 mg, 5 mg, 10 mg, 50 mg, 100 mg Amentoflavone ent-11β-Hydroxyatis-16-ene-3,14-dione (Didemethyl-ginkgetin) Cat. -

Study Initiation Q4 2017 - 18
Study Initiation Q4 2017 - 18 Reasons for Research Ethics Integrated Research Date Site Non- Date of First Date Site . Date Site HRA Approval Date Site Date Site Ready Benchmark delay Committee Application System Name of Trial Confirmed By Confirmation Patient Reasons for Delay Comments Invited Selected Date Confirmed To Start Met correspond Reference Number Number Sponsor Status Recruited to: HRA pack was received prematurely PLATO - PersonaLising Anal cancer radioTherapy dOse - 16/YH/0157 204585 21/07/2016 12/04/2017 20/07/2016 30/06/2017 13/06/2017 Please Select... 14/08/2017 21/06/2017 Yes A - Permissions delayed/denied from sponsor. Delays with site capacity Both Incorporating Anal Cancer Trials (ACT) ACT3, ACT4 and ACT5 and capability review. MAnagement of high bleeding risk patients post bioresorbable A - Site documentation delays. Rare 17/LO/0108 221119 polymer coated STEnt implantation with an abbReviated versus 23/12/2016 10/04/2017 10/04/2017 29/03/2017 04/04/2017 Please Select... 30/05/2017 01/08/2017 No A - Permissions delayed/denied NHS Provider patient group prolonged DAPT regimen – MASTER DAPT Delays by sponsor - portfolio adoption awaited and then site pharmacy concerns remained unresolved by Cereal Bar or oral supplementation with tablets to increase serum Site declined to Site Not 16/LO/1130 187152 01/02/2017 05/05/2017 01/11/2016 D - Sponsor Delays sponsor and it was decided to close Sponsor folate in young pregnant women participate Confirmed BSUH as a site on the 17/08/17, therefore we did not open as a site for this study 70 Day target date missed due to sponsor delays with devices and delays with MHRA & HRA Approvals. -

A Meeting Report from the 6Th Isirv Antiviral Group Conference T
Antiviral Research 167 (2019) 45–67 Contents lists available at ScienceDirect Antiviral Research journal homepage: www.elsevier.com/locate/antiviral Advances in respiratory virus therapeutics – A meeting report from the 6th isirv Antiviral Group conference T ∗ John H. Beigela, , Hannah H. Namb, Peter L. Adamsc, Amy Kraffta, William L. Inced, Samer S. El-Kamaryd, Amy C. Simse a National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA bNorthwestern University, Feinberg School of Medicine, Chicago, IL, USA c Biomedical Advanced Research and Development Authority (BARDA), Office of the Assistant Secretary for Preparedness and Response (ASPR), Department of Health and Human Services (HHS), Washington, DC, USA d Division of Antiviral Products, Office of Antimicrobial Products, Office of New Drugs, Center for Drug Evaluation and Research, U.S Food and Drug Administration, Silver Spring, MD, USA e Gillings School of Global Public Health, Department of Epidemiology, University of North Carolina, Chapel Hill, NC, USA ARTICLE INFO ABSTRACT Keywords: The International Society for Influenza and other Respiratory Virus Diseases held its 6th Antiviral Group (isirv- Influenza AVG) conference in Rockville, Maryland, November 13–15, 2018. The three-day program was focused on Respiratory syncytial virus therapeutics towards seasonal and pandemic influenza, respiratory syncytial virus, coronaviruses including Coronavirus MERS-CoV and SARS-CoV, human rhinovirus, and other respiratory viruses. Updates were presented on several Antiviral therapy influenza antivirals including baloxavir, CC-42344, VIS410, immunoglobulin, immune plasma, MHAA4549A, Host-directed therapeutics pimodivir (JNJ-63623872), umifenovir, and HA minibinders; RSV antivirals including presatovir (GS-5806), ziresovir (AK0529), lumicitabine (ALS-008176), JNJ-53718678, JNJ-64417184, and EDP-938; broad spectrum antivirals such as favipiravir, VH244, remdesivir, and EIDD-1931/EIDD-2801; and host directed strategies in- cluding nitazoxanide, eritoran, and diltiazem. -

Stembook 2018.Pdf
The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 WHO/EMP/RHT/TSN/2018.1 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances. Geneva: World Health Organization; 2018 (WHO/EMP/RHT/TSN/2018.1). Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -

Elucidation of the Roles of PTN and ISG15 in RSV Cytopathogenesis: Possible Biomarkers of Severe Disease?
DOCTOR OF PHILOSOPHY Elucidation of the roles of PTN and ISG15 in RSV cytopathogenesis: possible biomarkers of severe disease? Groves, Helen Award date: 2018 Awarding institution: Queen's University Belfast Link to publication Terms of use All those accessing thesis content in Queen’s University Belfast Research Portal are subject to the following terms and conditions of use • Copyright is subject to the Copyright, Designs and Patent Act 1988, or as modified by any successor legislation • Copyright and moral rights for thesis content are retained by the author and/or other copyright owners • A copy of a thesis may be downloaded for personal non-commercial research/study without the need for permission or charge • Distribution or reproduction of thesis content in any format is not permitted without the permission of the copyright holder • When citing this work, full bibliographic details should be supplied, including the author, title, awarding institution and date of thesis Take down policy A thesis can be removed from the Research Portal if there has been a breach of copyright, or a similarly robust reason. If you believe this document breaches copyright, or there is sufficient cause to take down, please contact us, citing details. Email: [email protected] Supplementary materials Where possible, we endeavour to provide supplementary materials to theses. This may include video, audio and other types of files. We endeavour to capture all content and upload as part of the Pure record for each thesis. Note, it may not be possible in all instances to convert analogue formats to usable digital formats for some supplementary materials. -

Review : Imunoterapi Penanganan Infeksi Virus
Jurnal Mandala Pharmacon Indonesia, Vol 7.No.1 Juni 2021 Avaiable online at www.jurnal-pharmaconmw.com/jmpi p-ISSN : 2442-6032 e-ISSN : 2598-9979 Review : Imunoterapi Penanganan Infeksi Virus Usmar1, Andi Maqhfirah Nurul Fitri1, Dewi Yuliana2, Firzan Nainu1* 1Fakultas Farmasi, Universitas Hasanuddin, Makassar 2Fakultas Farmasi, Universitas Muslim Indonesia, Makassar ABSTRAK Penyakit menular akibat virus merupakan salah satu perkembangan yang pesat terutama dalam kondisi masalah kesehatan global yang mempengaruhi sistem pandemi COVID-19 yang dihadapi saat ini. Meskipun kesehatan masyarakat dan ekonomi di seluruh dunia. terapi dan obat-obatan yang digunakan dalam bidang Hal tersebut mendorong disusunnya artikel ini untuk imunofarmakologi masih terbatas serta banyak hal yang mendiskusikan relevansi dan pentingnya imunoterapi belum dapat ditemukan, namun teknologi baru dan dalam menentukan pilihan terapetik terkait infeksi kemajuan pesat dalam pengetahuan tentang regulasi virus, dengan menitikberatkan pembahasan pada sistem imun telah menjadikan imunoterapi sebagai ketersediaan pilihan vaksin dan obat yang telah bidang yang memiliki potensi besar dan menjanjikan ditemukan untuk membantu manusia melawan berbagai dalam penanganan infeksi virus maupun patogen lain. jenis infeksi yang disebabkan oleh virus. Penulisan Oleh karena itu, konsep imunoterapi serta relevansinya artikel review naratif ini dilakukan menggunakan dengan penyakit manusia merupakan salah satu solusi metode analisis pustaka primer maupun sekunder yang menawarkan pilihan baru -

United States Bankruptcy Court for the District of Delaware
Case 17-10805-LSS Doc 409 Filed 11/02/17 Page 1 of 268 IN THE UNITED STATES BANKRUPTCY COURT FOR THE DISTRICT OF DELAWARE In re: Chapter 11 UNILIFE CORPORATION, et al., 1 Case No. 17-10805 (LSS) Debtors. (Jointly Administered) AFFIDAVIT OF SERVICE STATE OF CALIFORNIA } } ss.: COUNTY OF LOS ANGELES } Darleen Sahagun, being duly sworn, deposes and says: 1. I am employed by Rust Consulting/Omni Bankruptcy, located at 5955 DeSoto Avenue, Suite 100, Woodland Hills, CA 91367. I am over the age of eighteen years and am not a party to the above- captioned action. 2. On October 30, 2017, I caused to be served the: Notice/Debtors’ Motion for Approval of Settlement of Certain Claims with Present and Former Officers and Directors, and Certain Plaintiffs and Their Counsel [Docket No. 406] Notice of Filing of Corrected Exhibit [Docket No. 407] By causing true and correct copies to be served via first-class mail, postage pre-paid to the names and addresses of the parties listed as follows: I. Docket No. 406 and Docket No. 407 to those parties listed on the annexed Exhibit A, II. Docket No. 406 (Notice Only) to those parties on the annexed Exhibit B, Also, by causing true and correct copies to be served via email to the parties listed as follows: /// 1 The Debtors in these chapter 11 cases are the following entities (the last four digits of each Debtor’s respective federal tax identification number, if any, follow in parentheses): Unilife Corporation (9354), Unilife Medical Solutions, Inc. (9944), and Unilife Cross Farm LLC (3994).