Genome-Wide Association Study Reveals Genetic Link Between
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dynamic Interplay Between Locus-Specific DNA Methylation and Hydroxymethylation Regulates Distinct Biological Pathways in Prostate Carcinogenesis Shivani N
Kamdar et al. Clinical Epigenetics (2016) 8:32 DOI 10.1186/s13148-016-0195-4 RESEARCH Open Access Dynamic interplay between locus-specific DNA methylation and hydroxymethylation regulates distinct biological pathways in prostate carcinogenesis Shivani N. Kamdar1,2, Linh T. Ho1, Ken J. Kron1, Ruth Isserlin3, Theodorus van der Kwast1,4, Alexandre R. Zlotta5, Neil E. Fleshner6, Gary Bader3 and Bharati Bapat1,2* Abstract Background: Despite the significant global loss of DNA hydroxymethylation marks in prostate cancer tissues, the locus-specific role of hydroxymethylation in prostate tumorigenesis is unknown. We characterized hydroxymethylation and methylation marks by performing whole-genome next-generation sequencing in representative normal and prostate cancer-derived cell lines in order to determine functional pathways and key genes regulated by these epigenomic modifications in cancer. Results: Our cell line model shows disruption of hydroxymethylation distribution in cancer, with global loss and highly specific gain in promoter and CpG island regions. Significantly, we observed locus-specific retention of hydroxymethylation marks in specific intronic and intergenic regions which may play a novel role in the regulation of gene expression in critical functional pathways, such as BARD1 signaling and steroid hormone receptor signaling in cancer. We confirm a modest correlation of hydroxymethylation with expression in intragenic regions in prostate cancer, while identifying an original role for intergenic hydroxymethylation in differentially expressed regulatory pathways in cancer. We also demonstrate a successful strategy for the identification and validation of key candidate genes from differentially regulated biological pathways in prostate cancer. Conclusions: Our results indicate a distinct function for aberrant hydroxymethylation within each genomic feature in cancer, suggesting a specific and complex role for the deregulation of hydroxymethylation in tumorigenesis, similar to methylation. -

DDB1 Functions As a Linker to Recruit Receptor WD40 Proteins to CUL4− ROC1 Ubiquitin Ligases
Downloaded from genesdev.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press RESEARCH COMMUNICATION domain to bind with a conserved protein motif, the F DDB1 functions as a linker box, which, via its additional protein–protein interaction to recruit receptor WD40 modules, recruits various substrates, often phosphory- lated, to the CUL1–ROC1 catalytic core. To bring spe- proteins to CUL4–ROC1 cific substrates to CUL2- and CUL5-dependent ligases, a ubiquitin ligases heterodimeric linker complex containing elongins B and C binds simultaneously to an analogous N-terminal do- Yizhou Joseph He, Chad M. McCall, Jian Hu, main in CUL2 and CUL5 and to two similar protein 1 motifs, the VHL box and SOCS box. VHL and SOCS pro- Yaxue Zeng, and Yue Xiong teins, via their additional protein–protein interaction Lineberger Comprehensive Cancer Center, Department of modules, target various substrates differentially to the Biochemistry and Biophysics, Program in Molecular Biology CUL2–ROC1 or CUL5–ROC2 catalytic cores (Kamura et and Biotechnology, University of North Carolina, Chapel Hill, al. 1998, 2001, 2004; Stebbins et al. 1999; Zhang et al. North Carolina 27599, USA 1999). Omitting a linker, CUL3 utilizes its N-terminal domain to bind to proteins with a conserved 100-residue Cullins assemble the largest family of ubiquitin ligases protein motif known as a BTB domain, which, via addi- by binding with ROC1 and various substrate receptors. tional protein–protein interaction domains, then target CUL4 function is linked with many cellular processes, various substrates to the CUL3–ROC1 catalytic core (Fu- rukawa et al. 2003; Geyer et al. -

Expression and Purification of Functional Recombinant CUL2
www.nature.com/scientificreports OPEN Expression and purifcation of functional recombinant CUL2•RBX1 from E. coli Stephanie Diaz1, Lihong Li1,2, Kankan Wang1 & Xing Liu1,2* Cullin-2 (CUL2) based cullin-RING ligases (CRL2s) comprise a family of ubiquitin E3 ligases that exist only in multi-cellular organisms and are crucial for cellular processes such as embryogenesis and viral pathogenesis. CUL2 is the scafold protein that binds one of the interchangeable substrate receptor modules, which consists of adaptor proteins and the substrate receptor protein. The VHL protein is a substrate receptor known to target hypoxia-inducible factor α (HIF1α) for ubiquitination and degradation. Because of its critical role in the ubiquitination of important cellular factors such as HIF1α, CRL2s have been investigated for their biological functions and the development of novel therapeutics against diseases. Given the importance of CRL2s in biological and biomedical research, methods that efciently produce functional CUL2 proteins will greatly facilitate studies on the mechanism and regulation of CRL2s. Here, we report two cost-efective systems for the expression and purifcation of recombinant human CUL2 from E. coli cells. The purifed CUL2 proteins were ~ 95% pure, could bind their substrate receptor modules, and were enzymatically active in transferring ubiquitin or ubiquitin-like protein to the corresponding substrate in in vitro assays. The presented methodological advancements will help advance research in CRL2 function and regulation. Protein turnover is a cellular regulatory system defned by the continuous synthesis and decomposition of specifc proteins to maintain the integrity of optimally functioning proteins 1,2. Abnormalities during protein turnover, specifcally during protein degradation, ofen result in human diseases such as cystic fbrosis and liposarcoma. -

The Structure and Regulation of Cullin 2 Based E3 Ubiquitin Ligases and Their Biological Functions Weijia Cai* and Haifeng Yang*
Cai and Yang Cell Div (2016) 11:7 DOI 10.1186/s13008-016-0020-7 Cell Division REVIEW Open Access The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions Weijia Cai* and Haifeng Yang* Abstract Background: Cullin-RING E3 ubiquitin ligase complexes play a central role in targeting cellular proteins for ubiquit- ination-dependent protein turnover through 26S proteasome. Cullin-2 is a member of the Cullin family, and it serves as a scaffold protein for Elongin B and C, Rbx1 and various substrate recognition receptors to form E3 ubiquitin ligases. Main body of the abstract: First, the composition, structure and the regulation of Cullin-2 based E3 ubiquitin ligases were introduced. Then the targets, the biological functions of complexes that use VHL, Lrr-1, Fem1b, Prame, Zyg-11, BAF250, Rack1 as substrate targeting subunits were described, and their involvement in diseases was discussed. A small molecule inhibitor of Cullins as a potential anti-cancer drug was introduced. Furthermore, proteins with VHL box that might bind to Cullin-2 were described. Finally, how different viral proteins form E3 ubiquitin ligase complexes with Cullin-2 to counter host viral defense were explained. Conclusions: Cullin-2 based E3 ubiquitin ligases, using many different substrate recognition receptors, recognize a number of substrates and regulate their protein stability. These complexes play critical roles in biological processes and diseases such as cancer, germline differentiation and viral defense. Through the better understanding of their biology, we can devise and develop new therapeutic strategies to treat cancers, inherited diseases and viral infections. -

Modeling Genomic Diversity and Tumor Dependency in Malignant Melanoma
Research Article Modeling Genomic Diversity and Tumor Dependency in Malignant Melanoma William M. Lin,1,3,5 Alissa C. Baker,1,3 Rameen Beroukhim,1,3,5 Wendy Winckler,1,3,5 Whei Feng,1,3,5 Jennifer M. Marmion,7 Elisabeth Laine,8 Heidi Greulich,1,3,5 Hsiuyi Tseng,1,3 Casey Gates,5 F. Stephen Hodi,1 Glenn Dranoff,1 William R. Sellers,1,6 Roman K. Thomas,9,10 Matthew Meyerson,1,3,4,5 Todd R. Golub,2,3,5 Reinhard Dummer,8 Meenhard Herlyn,7 Gad Getz,3,5 and Levi A. Garraway1,3,5 Departments of 1Medical Oncology and 2Pediatric Oncology and 3Center for Cancer Genome Discovery, Dana-Farber Cancer Institute, Harvard Medical School; 4Department of Pathology, Harvard Medical School, Boston, Massachusetts; 5The Broad Institute of M.I.T. and Harvard; 6Novartis Institutes for Biomedical Research, Cambridge, Massachusetts; 7Cancer Biology Division, Wistar Institute, Philadelphia, Pennsylvania; 8Department of Dermatology, University of Zurich Hospital, Zu¨rich, Switzerland; 9Max Planck Institute for Neurological Research with Klaus Joachim Zulch Laboratories of the Max Planck Society and the Medical Faculty of the University of Cologne; and 10Center for Integrated Oncology and Department I for Internal Medicine, University of Cologne, Cologne, Germany Abstract tumorigenesis have been derived from functional studies involving The classification of human tumors based on molecular cultured human cancer cells (e.g., established cell lines, short-term cultures, etc.). Despite their limitations, cancer cell line collections criteria offers tremendous clinical potential; however, dis- cerning critical and ‘‘druggable’’ effectors on a large scale will whose genetic alterations reflect their primary tumor counterparts also require robust experimental models reflective of tumor should provide malleable proxies that facilitate mechanistic genomic diversity. -

Short Article CAND1 Binds to Unneddylated CUL1 and Regulates
Molecular Cell, Vol. 10, 1519–1526, December, 2002, Copyright 2002 by Cell Press CAND1 Binds to Unneddylated CUL1 Short Article and Regulates the Formation of SCF Ubiquitin E3 Ligase Complex Jianyu Zheng,1 Xiaoming Yang,1 Despite the importance of cullins in controlling many Jennifer M. Harrell,1 Sophia Ryzhikov,1 essential biological processes, the mechanism that reg- Eun-Hee Shim,1 Karin Lykke-Andersen,2 ulates the cullin-containing ubiquitin E3 ligases remains Ning Wei,2 Hong Sun,1 Ryuji Kobayashi,3 unclear. In SCF, the F box proteins are short-lived pro- and Hui Zhang1,4 teins that undergo CUL1/SKP1-dependent degradation 1Department of Genetics (Wirbelauer et al., 2000; Zhou and Howley, 1998). Dele- Yale University School of Medicine tion of the F box region abolishes the binding of F box 333 Cedar Street proteins to SKP1 and CUL1, and consequently increases 2 Department of Molecular, Cellular, the stability of F box proteins. This substrate-indepen- and Developmental Biology dent proteolysis of F box proteins is likely the result of Yale University autoubiquitination by the ubiquitin E2 and E1 enzymes New Haven, Connecticut 06520 through a CUL1/SKP1-dependent mechanism. 3 Cold Spring Harbor Laboratory The carboxy-terminal ends of cullins are often covalently Cold Spring Harbor, New York 11724 modified by a ubiquitin-like protein, NEDD8/RUB1, and this modification appears to associate with active E3 li- gases (Hochstrasser, 2000). Like ubiquitin modification, Summary neddylation requires E1 (APP-BP1 and UBA3)-activating and E2 (UBC12)-conjugating enzymes (Hochstrasser, The SCF ubiquitin E3 ligase regulates ubiquitin-depen- 2000). -
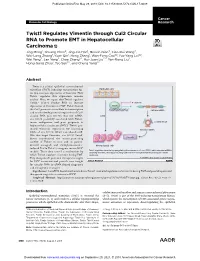
Twist1 Regulates Vimentin Through Cul2 Circular RNA to Promote EMT
Published OnlineFirst May 29, 2018; DOI: 10.1158/0008-5472.CAN-17-3009 Cancer Molecular Cell Biology Research Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma Jing Meng1, Shuang Chen2, Jing-Xia Han1, Baoxin Qian3, Xiao-Rui Wang4, Wei-Long Zhong1, Yuan Qin1, Heng Zhang1, Wan-Feng Gao1,2, Yue-Yang Lei1,2, Wei Yang2, Lan Yang2, Chao Zhang1,2, Hui-Juan Liu2,4, Yan-Rong Liu2, Hong-Gang Zhou1, Tao Sun1,2, and Cheng Yang1,2 Abstract Twist is a critical epithelial–mesenchymal transition (EMT)–inducing transcription fac- Epithelial cells tor that increases expression of vimentin. How Twist1 regulates this expression remains unclear. Here, we report that Twist1 regulates Cullin2 (Cul2) circular RNA to increase E-cadherin expressionofvimentininEMT.Twist1bound Twist1 theCul2promotertoactivateitstranscription Cul2 pre-mRNA and to selectively promote expression of Cul2 circular RNA (circ-10720), but not mRNA. EMT circ-10720 positively correlated with Twist1, tumor malignance, and poor prognosis in circRNA-10720 hepatocellular carcinoma (HCC). Twist1 pro- moted vimentin expression by increasing Vimentin miRNA levels of circ-10720, which can absorb miR- NAs that target vimentin. circ-10720 knock- down counteracted the tumor-promoting Vimentin activity of Twist1 in vitro and in patient- derived xenograft and diethylnitrosamine- Mesenchymal cells induced TetOn-Twist1 transgenic mouse HCC Twist1 regulates vimentin by upregulating the expression of circ-10720, which absorbs miRNAs models. These data unveil a mechanism by targeting vimentin, thereby promoting epithelial-mesenchymal transition in hepatocellular which Twist1 regulates vimentin during EMT. carcinoma. They also provide potential therapeutic targets © 2018 American Association for Cancer Research for HCC treatment and provide new insight forcircularRNA(circRNA)-baseddiagnostic and therapeutic strategies. -

Supporting Information
Supporting Information Friedman et al. 10.1073/pnas.0812446106 SI Results and Discussion intronic miR genes in these protein-coding genes. Because in General Phenotype of Dicer-PCKO Mice. Dicer-PCKO mice had many many cases the exact borders of the protein-coding genes are defects in additional to inner ear defects. Many of them died unknown, we searched for miR genes up to 10 kb from the around birth, and although they were born at a similar size to hosting-gene ends. Out of the 488 mouse miR genes included in their littermate heterozygote siblings, after a few weeks the miRBase release 12.0, 192 mouse miR genes were found as surviving mutants were smaller than their heterozygote siblings located inside (distance 0) or in the vicinity of the protein-coding (see Fig. 1A) and exhibited typical defects, which enabled their genes that are expressed in the P2 cochlear and vestibular SE identification even before genotyping, including typical alopecia (Table S2). Some coding genes include huge clusters of miRNAs (in particular on the nape of the neck), partially closed eyelids (e.g., Sfmbt2). Other genes listed in Table S2 as coding genes are [supporting information (SI) Fig. S1 A and C], eye defects, and actually predicted, as their transcript was detected in cells, but weakness of the rear legs that were twisted backwards (data not the predicted encoded protein has not been identified yet, and shown). However, while all of the mutant mice tested exhibited some of them may be noncoding RNAs. Only a single protein- similar deafness and stereocilia malformation in inner ear HCs, coding gene that is differentially expressed in the cochlear and other defects were variable in their severity. -

Structural Basis of Cullin-2 RING E3 Ligase Regulation by the COP9 Signalosome
bioRxiv preprint doi: https://doi.org/10.1101/483024; this version posted November 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome One sentence summary: Structure and dynamics of the CSN-CRL2 complexes assessed by cryo-electron microscopy and structural mass spectrometry. Sarah V. Faull1#, Andy. M. C. Lau2#, Chloe Martens2, Zainab Ahdash2, Hugo Yebenes1,3, Carla Schmidt4, Fabienne Beuron1, Nora B. Cronin1, Edward P. Morris1*, Argyris Politis2*& 1 Division of Structural Biology, The Institute of Cancer Research, London, SW3 6JB, UK 2 Department of Chemistry, King’s College London, 7 Trinity Street, London, SE1 1DB, UK 3 Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas, Madrid, Spain 4 Interdisciplinary Research Center HALOmem, Charles Tanford Protein Centre, Martin Luther University Halle-Wittenberg, Kurt-Mothes-Strasse 3a, 06120 Halle/Saale, Germany # These authors contributed equally to this work & Lead Contact * Correspondence: Edward Morris Tel: +44 (0) 20 7153 5531 Email: [email protected] Argyris Politis Tel: +44 (0) 20 7848 7514 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/483024; this version posted November 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. SUMMARY Cullin-Ring E3 Ligases (CRLs) regulate a multitude of cellular pathways through specific substrate receptors. The COP9 signalosome (CSN) deactivates CRLs by removing NEDD8 (N8) from activated Cullins. -

Gene Ontology Analysis of GWA Study Data Sets Provides Insights Into the Biology of Bipolar Disorder
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector ARTICLE Gene Ontology Analysis of GWA Study Data Sets Provides Insights into the Biology of Bipolar Disorder Peter Holmans,1,* Elaine K. Green,1 Jaspreet Singh Pahwa,1 Manuel A.R. Ferreira,2,3,4,6,7,8 Shaun M. Purcell,2,3,4,6,7 Pamela Sklar,2,3,4,5,6,7 The Wellcome Trust Case-Control Consortium,9 Michael J. Owen,1 Michael C. O’Donovan,1 and Nick Craddock1 We present a method for testing overrepresentation of biological pathways, indexed by gene-ontology terms, in lists of significant SNPs from genome-wide association studies. This method corrects for linkage disequilibrium between SNPs, variable gene size, and multiple testing of nonindependent pathways. The method was applied to the Wellcome Trust Case-Control Consortium Crohn disease (CD) data set. At a general level, the biological basis of CD is relatively well known for a complex genetic trait, and it thus acted as a test of the method. The method, known as ALIGATOR (Association LIst Go AnnoTatOR), successfully detected biological pathways implicated in CD. The method was also applied to a meta-analysis of bipolar disorder, and it implicated the modulation of transcription and cellular activity, including that which occurs via hormonal action, as an important player in pathogenesis. Introduction causing. To illustrate the application of the method, we defined groups on the basis of membership in Gene Genome-wide association (GWA) analysis can be a power- Ontology (GO) database categories, though the approach ful method for identifying genes involved in complex is applicable to any other gene-membership classification disorders, which often arise from the interplay of multiple system. -

Chromosome 10, Frequently Lost in Human Melanoma, Encodes Multiple Tumor-Suppressive Functions
Published OnlineFirst January 22, 2014; DOI: 10.1158/0008-5472.CAN-13-1446 Cancer Tumor and Stem Cell Biology Research Chromosome 10, Frequently Lost in Human Melanoma, Encodes Multiple Tumor-Suppressive Functions Lawrence N. Kwong and Lynda Chin Abstract Although many DNA aberrations in melanoma have been well characterized, including focal amplification and deletions of oncogenes and tumor suppressors, broad regions of chromosomal gain and loss are less well understood. One possibility is that these broad events are a consequence of collateral damage from targeting single loci. Another possibility is that the loss of large regions permits the simultaneous repression of multiple tumor suppressors by broadly decreasing the resident gene dosage and expression. Here, we test this hypothesis in a targeted fashion using RNA interference to suppress multiple candidate residents in broad regions of loss. We find that loss of chromosome regions 6q, 10, and 11q21-ter is correlated with broadly decreased expression of most resident genes and that multiple resident genes impacted by broad regional loss of chromosome 10 are tumor suppressors capable of affecting tumor growth and/or invasion. We also provide additional functional support for Ablim1 as a novel tumor suppressor. Our results support the hypothesis that multiple cancer genes are targeted by regional chromosome copy number aberrations. Cancer Res; 74(6); 1–8. Ó2014 AACR. Introduction mas (7). In contrast, the observed approximately 60% of CDKN2A/B Increasingly high-resolution genomic studies have estab- deletions on chromosome 9p are focal. These data PTEN lished that recurrent focal deletions and amplifications in suggest that although is a major driver of chromosome cancer can selectively target specific oncogenes and tumor 10 loss, other genes may also be targeted for inactivation. -

3Q26 Amplifications in Cervical Squamous Carcinomas
Article 3q26 Amplifications in Cervical Squamous Carcinomas Ioannis A. Voutsadakis 1,2 1 Algoma District Cancer Program, Sault Area Hospital, Sault Ste. Marie, ON P6B 0A8, Canada; [email protected] 2 Section of Internal Medicine, Division of Clinical Sciences, Northern Ontario School of Medicine, Sudbury, ON P3E 2C6, Canada Abstract: Background: Squamous carcinomas of the uterine cervix often carry mutations of the gene encoding for the catalytic sub-unit of kinase PI3K, PIK3CA. The locus of this gene at chromosome 3q26 and neighboring loci are also commonly amplified. The landscape of 3q26-amplified cases have not been previously characterized in detail in cervical cancer. Methods: Published genomic data and associated clinical data from TCGA cervical cancer cohort were analyzed at cBioportal for amplifications in genes at 3q26. The clinical and molecular characteristics of the group of patients with 3q26 amplifications was compared with the group without 3q26 amplifications. Comparative prevalence of amplification and expression of genes at 3q26 in amplified squamous cervical cancer cases were surveyed as well as 3q26 amplifications in cervical cancer cell line databases. Results: Amplification of 3q26 locus is a prevalent molecular lesion in cervical squamous cell carcinomas encountered in about 15% of cases in TCGA cohort of 247 patients. Cancer-related genes commonly amplified from 3q26 include PIK3CA, TBL1XR1, DCUN1D1, SOX2, MECOM, PRKCI, and TERC. Amplified cases do not completely overlap with PIK3CA mutant cases. Differences exist between 3q26-amplified and non-amplified carcinomas in the frequency of mutations and frequency of other amplifications. Most commonly over-expressed genes in 3q26 amplified cases include PIK3CA, TBL1XR1, DCUN1D1, and less commonly SOX2 and PRKCI.