Title of Thesis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

St. Mary's Catholic Church Cemetery Record of Mt. Calvary Cemetery, Grand Rapids, MI
St. Mary's Catholic Church Cemetery record of Mt. Calvary Cemetery, Grand Rapids, MI. Last name First name Lot # Block Grave # Date of Burial Owner Comments Abram Joseph J. SE E 1 7.31.1950 Mary Abram Paid $75 Aug 1, 1950 Abram Mary SE E 2 1.18.1954 Mary Abram Paid $75 Aug 1, 1950 Absmeir Carl 30 E 1 10.18.1956 Absmeir C. Absmeir Marie 30 E 2 6.9.1955 Absmeir C. Absmeir Baby 144 C 1 4.13.1924 Absmeir, Elsie 1 grave Adams Baby 1 1 6.4.1942 Adams Row 3 Grave 5 Adrian Mary B. 54 C 1 8.12.1930 Adrian, Miss North East Quarter 3 graves Albers Louis 79 D 1 5.15.1928 Albers, Louis Albers Margaret 79 D 2 10.5.1937 Albers, Louis Albert Thersea 198 D 4 10.16.1956 Alberts, Thersea West Half Albert 1453 4th St. To last owner Parrett by Albert Margaret 188 1 3 11.2.1949 Leitelt, Joseph & Dora Albert, Margaret wiever (Noterized) of all other heirs Alberts Patricia 198 D 1 1.24.1931 Alberts, Thersea West Half E Alberts Katherine 262 1 1 11.12.1910 Alberts, Adam East Half Lot is full. Alberts Adam 262 1 2 5.3.1930 Alberts, Adam East Half Lot is full. Alberts Emma 262 1 3 10.3.1906 Alberts, Adam East Half Lot is full. Alden Jr. Charles Robert 328 D 1 2.24.1941 Alden, Chas. Robt. 1 Grave Aldrich Ella B. 79 3 2 7.12.1941 VanHoeven, Helen South Half Aleszkiewicz Frances 26 3 2 7.16.1937 Graczyk, Frank Aleszkiewicz Vincent 26 3 3 5.29.1929 Graczyk, Frank Alexzkrewicz Joseph S. -

Her Majesty the Queen PROGRAMME of the Sixty-Fourth Annual Mod
P. >wAel.2S^ An Comunn Gaidhealach Patron : Her Majesty The Queen PROGRAMME OF THE SIXTY FOURTH ANNUAL MOD Glasgow/2-6 October 1967 <4s> ©tan Meet'tfixmeo ★ For Complete Photographic Coverage Of The Mod ★ ON SALE IN GLASGOW ON THURSDAY THE NEWSPAPER HIGHLANDERS READ FOR NEWS OF THE HIGHLANDS A MATTER OF Balance There is an easier, and much more sensible way to maintain a good footing in money matters; open a Clydesdale Bank Current Account, with a cheque book of your own, and enjoy modern banking facilities. The Bank's up-to-date accountancy systems will show you a clear picture of your income/expenditure balance. The Manager of your nearest Branch of the Clydesdale Bank will be happy to explain the Bank's services in detail, including the Midland Bank Group £30 Cheque Card. Clydesdale Bank Ltd Ob HEAD OFFICE : 30 St Vincent Place Glasgow C 1 OVER 350 BRANCHES FROM THE SOLWAY TO SHETLAND — i — Air son a’ Phiob Mhor agus Aodach na Gaidhealtachd do n Bhuth GRAINGER agus CAMPBELL LTD. 1191-1193 SRAID EARRA-GHAIDHEAL GLASCHU, C.3. Telefon : Central 9103 AN COMUNN G AID HEAL AC H Patron: Her Majesty The Queen PROGRAMME of the Sixty-Fourth Annual Mod CONTENTS PAGE Programme ■ 5 Executive and Regional Councils • 7 Facal bho’n Cheann Suidhe 9 Traditional Gaelic Song II Time-table 15 Street Plan 23 Junior Section—Written Competitions 25 Tuesday—Junior Section—Oral Delivery 27 Vocal Music ... 43 Instrumental Music 57 Senior Section—Written Competitions 63 Wednesday—Senior Section and Vocal Music 64 Thursday—Senior Section — Oral Delivery 75 Vocal Music 79 Clarsach Friday—Senior Section—Vocal Music 86 Instrumental Music 90 Winners of the Premier Competitions 93 — 3 — TODAY’S MEN OF ACTION ARE YOU GETTING ENOUGH OUT OF LIFEP Enough adventure? Travel? Prospects? See what life in today’s Army offers you For further details apply to Army Careers Information Office 518 Sauchiehall Street, Glasgow, C.2 Tel. -

A Foundation for the Future
A FOUNDATION FOR THE FUTURE INVESTORS REPORT 2012–13 NORTHWESTERN UNIVERSITY Dear alumni and friends, As much as this is an Investors Report, it is also living proof that a passion for collaboration continues to define the Kellogg community. Your collective support has powered the forward movement of our ambitious strategic plan, fueled development of our cutting-edge curriculum, enabled our global thought leadership, and helped us attract the highest caliber of students and faculty—all key to solidifying our reputation among the world’s elite business schools. This year, you also helped set a new record for alumni support of Kellogg. Our applications and admissions numbers are up dramatically. We have outpaced our peer schools in career placements for new graduates. And we have broken ground on our new global hub. Your unwavering commitment to everything that Kellogg stands for helps make all that possible. Your continuing support keeps us on our trajectory to transform business education and practice to meet the challenges of the new economy. Thank you for investing in Kellogg today and securing the future for generations of courageous leaders to come. All the best, Sally Blount ’92, Dean 4 KELLOGG.NORTHWESTERN.EDU/INVEST contentS 6 Transforming Together 8 Early Investors 10 Kellogg Leadership Circle 13 Kellogg Investors Leaders Partners Innovators Activators Catalysts who gave $1,000 to $2,499 who gave up to $1,000 99 Corporate Affiliates 101 Kellogg Investors by Class Year 1929 1949 1962 1975 1988 2001 1934 1950 1963 1976 1989 2002 -

Annual Report 2012 Letter from Jonny
ANNUAL REPORT 2012 LETTER FROM JONNY Imerman Angels in 2012 can best be described in one word, IMPACT. The impact that we have on cancer survivors, fighters and caregivers is why we are here and why we do what we do. Our team is responsible for making personalized 1-on-1 connections for lives touched by cancer. In 2012, our services helped individuals in all fifty states and in over sixty countries scattered all over the globe. I am proud of the team we have developed and the positive impact we continue to achieve. The success of our program enables the organization to grow in all aspects as a whole. Everyone who joins Imerman Angels, whether a survivor, fighter, caregiver or supporter, will help us reach our goal that ensures no one ever fights cancer alone. I sincerely want to thank everyone from the bottom of my heart who has helped this organization grow and get to where it is today. We would not be here without you. Thank you to the mentor angels who share their stories. Thank you to the staff who makes this all possible. Thank you to the volunteers and donors for your time and generosity. Thank you to everyone else who believes in our mission. The impact we have in the cancer community is felt around the world and we are on the road to ensure no one faces cancer alone. With love and respect, Jonny Imerman LETTER FROM JOHN 2012 Was another EXCITING year for Imerman Angels! We saw our program grow to all-time high levels. -

Waterloo County Soldier Information Cards - World War II
Waterloo County Soldier Information Cards - World War II Residence [R] or Last Name First Name Rank Regiment/Battalion Hometown [H] A H.Q. Company, Highland Light Infantry of Ableson Albert Private Canada Galt [H] Ableson Gordon L. Stoker First Class Royal Canadian Navy Galt [H] Adam Kenneth F. Pilot Officer Royal Canadian Air Force Elmira [H], Preston [R] Adamos John Private Essex Scottish Regiment Kitchener Adams G. n/a Veterans Guard of Canada Galt [R] Adams Hector J. Gunner Royal Canadian Artillery Preston Adams Hugh B. Trooper Royal Canadian Armoured Corps Norwood [H] Adams William C. n/a Highland Light Infantry of Canada Preston [H] Adams Lawrence R. Private Quebec Royal Rifles Kitchener Addis Harold Private Western Ontario Regiment Preston [H] Agnew Archie Sergeant Hastings and Price Edward Regiment Galt [R] Aigner Frank Lance Corporal Highland Light Infantry of Canada Waterloo Airdrie Douglas Private n/a Elora Aitchinson Edward Flight Lieutenant Royal Canadian Air Force Elora Aitken George M. Private Irish Regiment of Canada Galt [H] Aitken W.T. "Bill" Second Lieutenant "C" Company, Highland Light Infantry of Canada Galt South Dumfries Township Aitkin George Captain Sault Ste. Marie and Sudbury Regiment [H] Aksim R.E. Captain Intelligence Corps Waterloo [R] Aksim Victor Captain Royal Canadian Corps of Signals Waterloo Albert Leo N. Private Highland Light Infantry of Canada Preston [H], Kitchener [R] Albrecht George Private Essex Scottish Regiment Milverton Alderman Frederick Corporal Highland Light Infantry of Canada Galt [R] Aldworth G. Louis Pilot Officer Royal Canadian Air Force n/a Alexander Clem Lance Corporal "D" Company, Highland Light Infantry of Canada Galt [R] Alexander Jack Paratrooper Paratroop Units Hespeler Alexander James Private Royal Canadian Infantry Corps Hespeler Alexander Thomas W. -
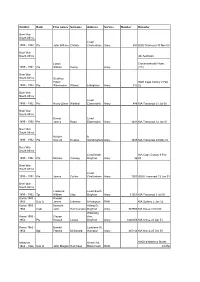
Conflict Rank First Names Surname Address Service Number Remarks
Conflict Rank First names Surname Address Service Number Remarks Boer War South Africa Lived 1899 - 1902 Pte John William Christie Cheltenham Army 401 DOS Transvaal 30 Nov 00 Boer War South Africa 4th Austalian Lytton Commonwealth Horse 1899 - 1902 Pte William Penny Army (Vic) Boer War South Africa Geoffrey Edwin DOS Cape Colony 8 Feb 1899 - 1902 Pte Winchester Wilmot b Brighton Army 339 02 Boer War South Africa Lived 1899 - 1902 Pte Henry Oliver Walford Elsternwick Army 488 KIA Transvaal 21 Jul 00 Boer War South Africa Ernest Lived 1899 - 1902 Pte James Rowe Elsternwick Army 1481 KIA Transvaal 12 Jun 01 Boer War South Africa Herbert b 1899 - 1902 Pte Vincent Hewins Sandringham Army 1455 KIA Transvaal 29 May 01 Boer War South Africa Lived North KIA Cape Colony 9 Feb 1899 - 1902 Pte Michael Conway Brighton Army 96 00 Boer War South Africa Lived 1899 - 1902 Pte James Collins Cheltenham Army 1007 DOW Transvaal 12 Jun 01 Boer War South Africa Frederick Lived South 1899 - 1902 Tpr William Clay Brighton Army 31561 KIA Transvaal 3 Jul 01 Korea 1950 - Ronald 1953 Sub Lt James Coleman b Hampton RAN KIA Sydney 2 Jan 52 Korea 1950 - Kenneth Asling St, 1953 Capt John Hummerston Brighton Army 337558 KIA Korea 3 Oct 50 Willansby Korea 1950 - Claude Ave, 1953 Pte Russell Looker Brighton Army 3400258 KIA Korea 24 Apr 51 Korea 1950 - Donald Ludstone St, 1953 Sgt Francis McDonald Hampton Army 345146 KIA Korea 25 Oct 50 Malaysia Beach Rd, A/Killed Malacca Straits 1964 - 1966 Sub Lt John Morgan Hutchison Black Rock RAN 24-Mar DOW South Vietnam 14 Dec Vietnam -

Surname Given Age Date Page Maiden Note Abildua Frank 91 22-Dec D-1 Ables Cary James 14 13-Apr D-1 Full Name Abrahamson Ethel 91 9-Mar D-2 Newman Abramson Rose H
Surname Given Age Date Page Maiden Note Abildua Frank 91 22-Dec D-1 Ables Cary James 14 13-Apr D-1 Full name Abrahamson Ethel 91 9-Mar D-2 Newman Abramson Rose H. 81 2-Feb D-2 Accordini Mary G. 70 20-Dec A-13 Ackerman Frances L. 4-May C-5 Adams Eva 83 3-Apr D-2 Stewart Adams Michael J. 76 28-Jan D-1 Adell Alfred S. 48 5-May B-4 Adoba William J., Sr. 64 10-Jan B-6 Agnew W. Lynn 85 12-Sep B-6 Albaugh James 81 9-Oct E-1 Albee Daniel 62 27-Jul D-1 Albert Henrietta 79 13-Apr D-1 Albright Ralph G. 80 13-Apr D-1 Aldrin Myrtle A. 76 15-Jan D-6 Alexander Daniel L. 37 13-Jun C-5 Alexander W. (Rev.) 69 6-Jan D-2 Alexich Jacov "Jack" 60 26-Dec D-1 Allande Arthur 80 13-Oct D-3 Allande Augustina 79 2-May C-2 Allen James P., Sr. 49 30-Oct D-1 Allen Ernest L. 48 6-Sep B-5 Veteran of the Korean conflict Allen Lizzie 3-Jan A-11 Allen William M. 75 21-Mar C-2 Allen Aryln J. 60 5-Jun F-7 Allen Annie 101 4-Feb D-1 Aller Edward D. 81 11-Jan D-2 Almanza Maria 81 13-Feb D-1 Aloia Frank, Sr. 90 9-Feb C-2 Alongi Samuel 70 5-Jan C-5 Veteran of World War II Alpert Samual 97 15-Dec C-7 Alsdorf Rose Evelyn 52 30-Sep C-3 Full name Alsip Yvetta E. -

April 30, 2016 | Michigan Stadium SPRING COMMENCEMENT UNIVERSITY of MICHIGAN April 30, 2016 10:00 A.M
April 30, 2016 | Michigan Stadium SPRING COMMENCEMENT UNIVERSITY OF MICHIGAN April 30, 2016 10:00 a.m. This program includes a list of the candidates for degrees to be granted upon completion of formal requirements. Candidates for graduate degrees are recommended jointly by the Executive Board of the Horace H. Rackham School of Graduate Studies and the faculty of the school or college awarding the degree. Following the School of Graduate Studies, schools are listed in order of their founding. Candidates within those schools are listed by degree then by specialization, if applicable. Horace H. Rackham School of Graduate Studies ..................................................................................................20 College of Literature, Science, and the Arts ............................................................................................................31 Medical School ......................................................................................................................................................51 Law School ............................................................................................................................................................52 School of Dentistry ................................................................................................................................................54 College of Pharmacy ..............................................................................................................................................55 -

Shrine Prayer Companion Ministry
January 5, 2020 | Epiphany of the Lord Shrine Herald NATIONAL SHRINE OF THE LITTLE FLOWER BASILICA ROMAN CATHOLIC CHURCH We Will Be A Band Of Joyful Missionary Disciples. On December 20, Archbishop Allen Vigneron announced the appointement of Reverend Joseph Horn as the next pastor of the National Shrine of the Little Flower Basilica, effective February 3, 2020. Mark your calendars: Fr. Horn will succeed Bishop-Elect Robert McClory, Ascend Page 14 who has been appointed by Pope Francis to serve begins January 22 as the bishop of the diocese of Gary, Indiana. Shrine Prayer Summit ~ February 29 No stranger to Shrine, Fr. Horn began his first Shrine Leadership Summit ~ March 28 See Page 8 for details assignment here as Associate Pastor from 1997- 2001 where he worked in youth ministry and the Life Teen Program. Ordained a priest in 1997 for the Archdiocese of Detroit, Fr. Horn went on to serve as Continued on page 7 2100 W. Twelve Mile Road, Royal Oak, MI 48073-3910 | (248) 541-4122 | www.shrinechurch.com Parish Office Information Our Weekly Homilies, Live! NATIONAL SHRINE of the LITTLE FLOWER BASILICA Access www.shrinechurch.com 2100 West Twelve Mile Road, Royal Oak, MI 48073.3910 Scroll down on the Home page to: PASTORAL STAFF/ PERSONAL PASTORAL Parish Main Number ......................................................................................... 248.541.4122 Bishop-Elect Robert McClory, Pastor-Rector ........................................................................ ext. 1415 Rev. Joseph Lang, Associate ........................................................................................................ -

Most Common Surnames
Most Common Surnames Surnames occurring most often in Scotland's registers of Births, Marriages and Deaths Counting only the surname of the child for births, the surnames of both bride and groom for marriages, and the surname of the deceased for deaths Note: the surnames from these registers may not be representative of the surnames of the population of Scotland as a whole, as (a) they include the surnames of non-residents who were born / married / died here; (b) they exclude the surnames of residents who were born / married / died elsewhere; and (c) some age-groups have very low birth, marriage and death rates; others account for most births, marriages and deaths. Registration Year=2015 Position Surname Number 1 SMITH 1929 2 BROWN 1438 3 WILSON 1352 4 STEWART 1186 5 THOMSON 1172 6 ROBERTSON 1158 7 CAMPBELL 1130 8 ANDERSON 1068 9 MURRAY 832 10 MACDONALD 823 11 TAYLOR 794 12 SCOTT 779 13 REID 771 14 CLARK 746 15 YOUNG 639 16 MORRISON 623 17 WALKER 619 18 ROSS 614 19 WATSON 595 20 GRAHAM 571 21 MITCHELL 566 22 FRASER 564 23 PATERSON 557 24 MILLER 555 25 HENDERSON 540 26 MARTIN 539 27 CAMERON 533 28 MCDONALD 531 29 HAMILTON 530 30= DAVIDSON 519 30= GRAY 519 32 DUNCAN 512 33= JOHNSTON 501 33= KERR 501 33= SIMPSON 501 36 HUNTER 477 37 BELL 474 38 FERGUSON 465 39 KELLY 459 40 ALLAN 442 41 GRANT 429 42 MACLEOD 425 43 MCLEAN 411 44 BLACK 404 45 MACKAY 402 46 WRIGHT 393 47 MACKENZIE 385 48 GIBSON 382 49 MARSHALL 379 50 KENNEDY 377 © Crown Copyright 2016 Most Common Surnames (continued) 51 WALLACE 371 52 JONES 368 53 RUSSELL 358 54 SUTHERLAND 351 -

November 2020 Vol 26, No 11, November 2020
® November 2020 1916. Oil on canvas, National 39.7 Museumsin x 62.5 in/101Liverpool, cm Liverpool,x 159 Art cm. Gallery, Lever UK. Lady A Tale from The Decameron, A Tale Respiratory Infections Respiratory John William (1849–1917), Waterhouse Vol 26, No 11, November 2020 DEPARTMENT OF HEALTH & HUMAN SERVICES Public Health Service Centers for Disease Control and Prevention (CDC) Mailstop D61, Atlanta, GA 30329-4027 Official Business Penalty for Private Use $300 Return Service Requested ISSN 1080-6040 Peer-Reviewed Journal Tracking and Analyzing Disease Trends Pages 2543–2796 ® EDITOR-IN-CHIEF D. Peter Drotman ASSOCIATE EDITORS EDITORIAL BOARD Charles Ben Beard, Fort Collins, Colorado, USA Barry J. Beaty, Fort Collins, Colorado, USA Ermias Belay, Atlanta, Georgia, USA Martin J. Blaser, New York, New York, USA David M. Bell, Atlanta, Georgia, USA Andrea Boggild, Toronto, Ontario, Canada Sharon Bloom, Atlanta, Georgia, USA Christopher Braden, Atlanta, Georgia, USA Richard Bradbury, Melbourne, Australia Arturo Casadevall, New York, New York, USA Mary Brandt, Atlanta, Georgia, USA Kenneth G. Castro, Atlanta, Georgia, USA Corrie Brown, Athens, Georgia, USA Vincent Deubel, Shanghai, China Charles H. Calisher, Fort Collins, Colorado, USA Christian Drosten, Charité Berlin, Germany Benjamin J. Cowling, Hong Kong, China Anthony Fiore, Atlanta, Georgia, USA Michel Drancourt, Marseille, France Isaac Chun-Hai Fung, Statesboro, Georgia, USA Paul V. Effler, Perth, Australia Kathleen Gensheimer, College Park, Maryland, USA David O. Freedman, Birmingham, Alabama, USA Rachel Gorwitz, Atlanta, Georgia, USA Peter Gerner-Smidt, Atlanta, Georgia, USA Duane J. Gubler, Singapore Stephen Hadler, Atlanta, Georgia, USA Richard L. Guerrant, Charlottesville, Virginia, USA Matthew J. Kuehnert, Edison, New Jersey, USA Scott Halstead, Arlington, Virginia, USA Nina Marano, Atlanta, Georgia, USA David L. -

This Marks the Connection of British Columbia's Section of the Great
The Great Trail in British Columbia Le Grand Sentier au Colombie-Britannique This marks the connection of British Columbia’s section of The Great Trail of Canada in honour of Canada’s 150th Ceci marque le raccordement du Grand Sentier à travers Colombie-Britannique pour le 150e anniversaire de la anniversary of Confederation in 2017. Confédération canadienne en 2017. À partir d’où vous êtes, vous pouvez entreprendre l’un des voyages les plus beaux et les plus diversifiés du monde. From where you are standing, you can embark upon one of the most magnificent and diverse journeys in the world. Que vous vous dirigiez vers l’est, l’ouest, le nord ou le sud, Le Grand Sentier du Canada — créé par le sentier Whether heading east, west, north or south, The Great Trail—created by Trans Canada Trail (TCT) and its partners— Transcanadien (STC) et ses partenaires — vous offre ses multiples beautés naturelles ainsi que la riche histoire et offers all the natural beauty, rich history and enduring spirit of our land and its peoples. l’esprit qui perdure de notre pays et des gens qui l’habitent. Launched in 1992, just after Canada’s 125th anniversary of Confederation The Great Trail was conceived by a group of Lancé en 1992, juste après le 125e anniversaire de la Confédération du Canada, Le Grand Sentier a été conçu, par un visionary and patriotic individuals as a means to connect Canadians from coast to coast to coast. groupe de visionnaires et de patriotes, comme le moyen de relier les Canadiens d’un océan aux deux autres.