Differentiation and Establishment of Dental Epithelial-Like Stem Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Ono -- PTH-Pthrp Receptor Signaling in Osterix-Expressing Progenitors.Pdf
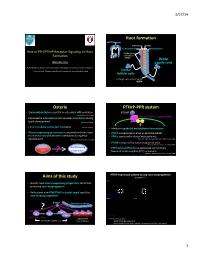
3/27/14 Root forma)on Cementum Dentin Cementoblast Odontoblast Role of PTH/PTHrP Receptor Signaling on Root Epithelial rests Formaon of Malassez (ERM) Dental Wanida Ono papilla cells AGE Orthodon;cs, Department of Developmental Biology, Harvard School of Dental Medicine Endocrine Unit, MassachuseLs General Hospital and Harvard Medical School Dental follicle cells Hertwig’s epithelial root sheath (HERS) Osterix PTHrP-PPR system • Transcripon factor essen;al to osteoblast differen;aon PTHrP (Nakashima K et al. 2002) PTH/PTHrP receptor (PPR) • Expressed in odontoblasts and alveolar osteoblasts during Gαs Gq tooth development (Chen S et al. 2009) • Controls cellular cementum formaon (Cao Z et al. 2012) • Mediates epithelial-mesenchymal interacons • Osterix-expressing precursors in the perichondrium move • PTHrP is expressed in enamel epithelia/HERS? to bone marrow and become osteoblasts during fetal • PPR is expressed in dental mesenchymes development (Maes C, Kronenberg HM et al. 2010) (Beck et al 1995; Lee Deeds and Segre 1995; Liu et al 1998) • PTHrP is required for tooth erup;on in mice (Philbrick WM, Karaplis AC et al. PNAS 1998) Osterix+ Root-forming • PPR haploinsufficiency is associated with primary cells progenitors failure of tooth erup;on (PFE) in humans ? (Decker E, Weber BH et al. Am J Hum Gen 2008) PTHrP expression paern during root morphogenesis Aims of this study PTHrPLacZ/+ x40 P7 P14 P49 • Iden;fy how osterix-expressing progenitors contribute to murine root morphogenesis • Understand how PTH/PTHrP receptor signal regulates root-forming progenitors PTHrP-LacZ x200 P7 x400 P14 ? PTHrP Epithelial root sheath PPR PTHrP-LacZ Osx+ progenitors Odontoblast PTHrP par;cipates in ……. -

Experimental Induction of Odontoblast Differentiation and Stimulation During Preparative Processes
Cells and Materials Volume 3 Number 2 Article 8 1993 Experimental Induction of Odontoblast Differentiation and Stimulation During Preparative Processes H. Lesot Institut de Biologie Médicale C. Begue-Kirn Institut de Biologie Médicale M. D. Kubler Institut de Biologie Médicale J. M. Meyer Institut de Biologie Médicale A. J. Smith Dental School, Birmingham See next page for additional authors Follow this and additional works at: https://digitalcommons.usu.edu/cellsandmaterials Part of the Biomedical Engineering and Bioengineering Commons Recommended Citation Lesot, H.; Begue-Kirn, C.; Kubler, M. D.; Meyer, J. M.; Smith, A. J.; Cassidy, N.; and Ruch, J. V. (1993) "Experimental Induction of Odontoblast Differentiation and Stimulation During Preparative Processes," Cells and Materials: Vol. 3 : No. 2 , Article 8. Available at: https://digitalcommons.usu.edu/cellsandmaterials/vol3/iss2/8 This Article is brought to you for free and open access by the Western Dairy Center at DigitalCommons@USU. It has been accepted for inclusion in Cells and Materials by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. Experimental Induction of Odontoblast Differentiation and Stimulation During Preparative Processes Authors H. Lesot, C. Begue-Kirn, M. D. Kubler, J. M. Meyer, A. J. Smith, N. Cassidy, and J. V. Ruch This article is available in Cells and Materials: https://digitalcommons.usu.edu/cellsandmaterials/vol3/iss2/8 Cells and Materials, Vol. 3, No. 2, 1993 (Pages201-217) 1051-6794/93$5. 00 +. 00 Scanning Microscopy International, Chicago (AMF O'Hare), IL 60666 USA EXPERIMENTAL INDUCTION OF ODONTOBLAST DIFFERENTIATION AND STIMULATION DURING REPARATIVE PROCESSES 1 1 1 2 2 1 H. -

Journal of Dental Research
Journal of Dental Research http://jdr.sagepub.com/ Cell Differentiation and Matrix Organization in Engineered Teeth A. Nait Lechguer, M.L. Couble, N. Labert, S. Kuchler-Bopp, L. Keller, H. Magloire, F. Bleicher and H. Lesot J DENT RES 2011 90: 583 originally published online 4 February 2011 DOI: 10.1177/0022034510391796 The online version of this article can be found at: http://jdr.sagepub.com/content/90/5/583 Published by: http://www.sagepublications.com On behalf of: International and American Associations for Dental Research Additional services and information for Journal of Dental Research can be found at: Email Alerts: http://jdr.sagepub.com/cgi/alerts Subscriptions: http://jdr.sagepub.com/subscriptions Reprints: http://www.sagepub.com/journalsReprints.nav Permissions: http://www.sagepub.com/journalsPermissions.nav >> Version of Record - Apr 13, 2011 OnlineFirst Version of Record - Feb 4, 2011 What is This? Downloaded from jdr.sagepub.com at Service Commun de la Documentation Université de Strasbourg on September 6, 2013 For personal use only. No other uses without permission. © 2011 International & American Associations for Dental Research RESEARCH REPORTS Biomaterials & Bioengineering A. Nait Lechguer1,2, M.L. Couble3,4, N. Labert3,4, S. Kuchler-Bopp1,2, Cell Differentiation and L. Keller1,2, H. Magloire3,4, F. Bleicher3,4, Matrix Organization in and H. Lesot1,2* Engineered Teeth 1INSERM UMR 977, Faculté de Médecine, 11, rue Humann, F-67085 Strasbourg, France; 2Dental School, University of Strasbourg, Strasbourg, France; 3Université de Lyon, Faculté d’Odontologie, Rue Guillaume Paradin, F-69372 Lyon Cedex 08, France; and 4IGFL, CNRS UMR 5242, Ecole Normale Supérieure, 46 Allée d’Italie, 69364, Lyon Cedex 08, France; *corresponding author, [email protected] J Dent Res 90(5):583-589, 2011 ABSTRACT InTRODuCTIOn Embryonic dental cells were used to check a series of criteria to be achieved for tooth engineering. -

Tooth Enamel and Its Dynamic Protein Matrix
International Journal of Molecular Sciences Review Tooth Enamel and Its Dynamic Protein Matrix Ana Gil-Bona 1,2,* and Felicitas B. Bidlack 1,2,* 1 The Forsyth Institute, Cambridge, MA 02142, USA 2 Department of Developmental Biology, Harvard School of Dental Medicine, Boston, MA 02115, USA * Correspondence: [email protected] (A.G.-B.); [email protected] (F.B.B.) Received: 26 May 2020; Accepted: 20 June 2020; Published: 23 June 2020 Abstract: Tooth enamel is the outer covering of tooth crowns, the hardest material in the mammalian body, yet fracture resistant. The extremely high content of 95 wt% calcium phosphate in healthy adult teeth is achieved through mineralization of a proteinaceous matrix that changes in abundance and composition. Enamel-specific proteins and proteases are known to be critical for proper enamel formation. Recent proteomics analyses revealed many other proteins with their roles in enamel formation yet to be unraveled. Although the exact protein composition of healthy tooth enamel is still unknown, it is apparent that compromised enamel deviates in amount and composition of its organic material. Why these differences affect both the mineralization process before tooth eruption and the properties of erupted teeth will become apparent as proteomics protocols are adjusted to the variability between species, tooth size, sample size and ephemeral organic content of forming teeth. This review summarizes the current knowledge and published proteomics data of healthy and diseased tooth enamel, including advancements in forensic applications and disease models in animals. A summary and discussion of the status quo highlights how recent proteomics findings advance our understating of the complexity and temporal changes of extracellular matrix composition during tooth enamel formation. -

6 Development of the Teeth: Root and Supporting Structures Nagat M
AVERY Chap.06 27-11-2002 10:09 Pagina 108 108 II Development of the Teeth and Supporting Structures 6 Development of the Teeth: Root and Supporting Structures Nagat M. ElNesr and James K. Avery Chapter Outline Introduction Introduction... 108 Objectives... 108 Root development is initiated through the contributions Root Sheath Development... 109 of the cells originating from the enamel organ, dental Single-Root Formation... 110 papilla, and dental follicle. The cells of the outer enamel Multiple-Root Formation... 111 epithelium contact the inner enamel epithelium at the Root Formation Anomalies... 112 base of the enamel organ, the cervical loop (Figs. 6.1 and Fate of the Epithelial Root Sheath (Hertwig's Sheath)... 113 6.2A). Later, with crown completion, the cells of the cer- Dental Follicle... 114 vical loop continue to grow away from the crown and Development of (Intermediate) Cementum... 116 become root sheath cells (Figs. 6.2B and 6.3). The inner Cellular and Acellular Cementum... 116 root sheath cells cause root formation by inducing the Development of the Periodontal Ligament... 117 adjacent cells of the dental papilla to become odonto- Development of the Alveolar Process... 119 blasts, which in turn will form root dentin. The root Summary... 121 sheath will further dictate whether the tooth will have Self-Evaluation Review... 122 single or multiple roots. The remainder of the cells of the dental papilla will then become the cells of the root pulp.The third compo- nent in root formation, the dental follicle, is the tissue that surrounds the enamel organ, the dental papilla, and the root. -

Dental Cementum Reviewed: Development, Structure, Composition, Regeneration and Potential Functions
Braz J Oral Sci. January/March 2005 - Vol.4 - Number 12 Dental cementum reviewed: development, structure, composition, regeneration and potential functions Patricia Furtado Gonçalves 1 Enilson Antonio Sallum 1 Abstract Antonio Wilson Sallum 1 This article reviews developmental and structural characteristics of Márcio Zaffalon Casati 1 cementum, a unique avascular mineralized tissue covering the root Sérgio de Toledo 1 surface that forms the interface between root dentin and periodontal Francisco Humberto Nociti Junior 1 ligament. Besides describing the types of cementum and 1 Dept. of Prosthodontics and Periodontics, cementogenesis, attention is given to recent advances in scientific Division of Periodontics, School of Dentistry understanding of the molecular and cellular aspects of the formation at Piracicaba - UNICAMP, Piracicaba, São and regeneration of cementum. The understanding of the mechanisms Paulo, Brazil. involved in the dynamic of this tissue should allow for the development of new treatment strategies concerning the approach of the root surface affected by periodontal disease and periodontal regeneration techniques. Received for publication: October 01, 2004 Key Words: Accepted: December 17, 2004 dental cementum, review Correspondence to: Francisco H. Nociti Jr. Av. Limeira 901 - Caixa Postal: 052 - CEP: 13414-903 - Piracicaba - S.P. - Brazil Tel: ++ 55 19 34125298 Fax: ++ 55 19 3412 5218 E-mail: [email protected] 651 Braz J Oral Sci. 4(12): 651-658 Dental cementum reviewed: development, structure, composition, regeneration and potential functions Introduction junction (Figure 1). The areas and location of acellular Cementum is an avascular mineralized tissue covering the afibrillar cementum vary from tooth to tooth and along the entire root surface. Due to its intermediary position, forming cementoenamel junction of the same tooth6-9. -

Amelogenic Transcriptome Profiling in Ameloblast-Like Cells Derived From
www.nature.com/scientificreports OPEN Amelogenic transcriptome profling in ameloblast-like cells derived from adult gingival epithelial cells Received: 23 May 2018 Sun-Yi Hyun1, Seyoung Mun1,2, Kyung-Jung Kang1, Jong-Chan Lim1, Shin-Young Kim1, Accepted: 29 January 2019 Kyudong Han1,2 & Young-Joo Jang 1 Published: xx xx xxxx Dental enamel is the highly mineralized tissue covering the tooth surface and is formed by ameloblasts. Ameloblasts have been known to be impossible to detect in adult tooth because they are shed by apoptosis during enamel maturation and tooth eruption. Owing to these, little was known about appropriate cell surface markers to isolate ameloblast-like cells in tissues. To overcome these problems, epithelial cells were selectively cultivated from the gingival tissues and used as a stem cell source for ameloblastic diferentiation. When gingival epithelial cells were treated with a specifed concentration of BMP2, BMP4, and TGFβ-1, the expression of ameloblast-specifc markers was increased, and both the MAPK and Smad signaling pathways were activated. Gingival epithelial cells diferentiated into ameloblast-like cells through epithelial-mesenchymal transition. By RNA-Seq analysis, we reported 20 ameloblast-specifc genes associated with cell surface, cell adhesion, and extracellular matrix function. These cell surface markers might be useful for the detection and isolation of ameloblast-like cells from dental tissues. Dentin, dental pulp, periodontal ligament, and dental enamel are developed by reciprocal interactions between dental epithelium and ectomesenchyme. Neural crest cell-derived ectomesenchyme diferentiates into odonto- blasts, periodontal ligament progenitors, cementoblasts, as well as various fibroblasts. On the other hand, enamel-forming ameloblasts diferentiate from epithelial cells originating from oral ectoderm. -

Basic Histology (23 Questions): Oral Histology (16 Questions
Board Question Breakdown (Anatomic Sciences section) The Anatomic Sciences portion of part I of the Dental Board exams consists of 100 test items. They are broken up into the following distribution: Gross Anatomy (50 questions): Head - 28 questions broken down in this fashion: - Oral cavity - 6 questions - Extraoral structures - 12 questions - Osteology - 6 questions - TMJ and muscles of mastication - 4 questions Neck - 5 questions Upper Limb - 3 questions Thoracic cavity - 5 questions Abdominopelvic cavity - 2 questions Neuroanatomy (CNS, ANS +) - 7 questions Basic Histology (23 questions): Ultrastructure (cell organelles) - 4 questions Basic tissues - 4 questions Bone, cartilage & joints - 3 questions Lymphatic & circulatory systems - 3 questions Endocrine system - 2 questions Respiratory system - 1 question Gastrointestinal system - 3 questions Genitouirinary systems - (reproductive & urinary) 2 questions Integument - 1 question Oral Histology (16 questions): Tooth & supporting structures - 9 questions Soft oral tissues (including dentin) - 5 questions Temporomandibular joint - 2 questions Developmental Biology (11 questions): Osteogenesis (bone formation) - 2 questions Tooth development, eruption & movement - 4 questions General embryology - 2 questions 2 National Board Part 1: Review questions for histology/oral histology (Answers follow at the end) 1. Normally most of the circulating white blood cells are a. basophilic leukocytes b. monocytes c. lymphocytes d. eosinophilic leukocytes e. neutrophilic leukocytes 2. Blood platelets are products of a. osteoclasts b. basophils c. red blood cells d. plasma cells e. megakaryocytes 3. Bacteria are frequently ingested by a. neutrophilic leukocytes b. basophilic leukocytes c. mast cells d. small lymphocytes e. fibrocytes 4. It is believed that worn out red cells are normally destroyed in the spleen by a. neutrophils b. -

Oral Histology
Oral Histology Lec-6 Dr. Nada AL.Ghaban Amelogenesis (Enamel formation) Amelogenesis begins at cusp tips and the incisal edges of the E.organ of the tooth germ and then it separated down the cusp slopes until all the cells of inner enamel epithelium(IEE) differentiate into ameloblasts. Amelogenesis begins shortly after dentinogenesis at the advanced or late bell stage. The delicate basement membrane between IEE and odontoblasts will disintegrate after dentinogenesis and before amelogenesis. During the early stages of tooth development, the IEE cells proliferate and contribute to the growth of the developing tooth. Ameloblasts fully differentiate at the growth centers located at cusp tips of the forming crown and this differentiation pattern spreads towards the cervical loop (the future cervical line in a fully formed tooth). However, once the IEE has fully differentiated into ameloblasts there is no more proliferation as these highly differentiated cells do not divide again. Amelogenesis is a complex process, it involves 2 stages which are: 1- E. matrix deposition. 2- Maturation or mineralization of the E. matrix. E. matrix deposition: It means the secretion of the E. matrix by ameloblasts. The freshly secreted E. matrix contain 30% minerals as hydroxy apatite crystals and 70% waters and E. proteins which include 90% amelogenine protein and 10% non-amelogenins protein( enameline and ameloblastin). These E. proteins which are secreted by ameloblasts are responsible for creating and 1 maintaining an extracellular environment favorable to mineral deposition. When the first layer of E. is laid down, the ameloblasts will begins to retreat from DEJ towards E. surface and begins to secrete the next layer of E. -

Ameloblastic Fibro-Odontoma of the Maxilla: a Case Report Belal Alani, Muraja Aldoori, Amar Adham, Farag Ismail
694 Case report Ameloblastic fibro-odontoma of the maxilla: a case report Belal Alani, Muraja Aldoori, Amar Adham, Farag Ismail HMC Hamad Medical Corporation, Doha, Qatar The ameloblastic fibro-odontoma is a rare benign odontogenic lesion defined as a Correspondence to Dr. Belal Alani, HMC tumor with the general features of ameloblastic fibroma but that also contains Hamad Medical Corporation, Doha, Qatar, enamel and dentin. In this article the authors describe a case of a young male PO Box 3050; patient with ameloblastic fibro-odontoma of the maxilla and the management of e-mail: [email protected] such condition. Received 8 April 2017 Accepted 3 June 2017 Keywords: The Egyptian Journal of Otolaryngology ameloblastic fibro-odontoma, ameloblastic, fibromaodontogenic tumor 2017, 33:694–697 Egypt J Otolaryngol 33:694–697 © 2017 The Egyptian Journal of Otolaryngology 1012-5574 Figure 1 Introduction The ameloblastic fibro-odontoma (AFO) is a rare benign odontogenic lesion defined as a tumor with the general features of ameloblastic fibroma but that also contains enamel and dentin [1]. According the recent WHO classification of odontogenic tumors published in 2005, AFO belongs to the group of lesions with odontogenic epithelium with odontogenic ectomesenchyme, with or without hard tissue formation [1]. AFO is normally found in young patients, with no significant sex predilection. The incidence of AFO is between 1 and 3% in odontogenic tumors [2,3]. Facial deformity and intraoral lesion with impingement on the left nostril. Clinically, it presents as a painless swelling of the affected area, usually the posterior portion of the Figure 2 maxilla or mandible. -

A Global Compendium of Oral Health
A Global Compendium of Oral Health A Global Compendium of Oral Health: Tooth Eruption and Hard Dental Tissue Anomalies Edited by Morenike Oluwatoyin Folayan A Global Compendium of Oral Health: Tooth Eruption and Hard Dental Tissue Anomalies Edited by Morenike Oluwatoyin Folayan This book first published 2019 Cambridge Scholars Publishing Lady Stephenson Library, Newcastle upon Tyne, NE6 2PA, UK British Library Cataloguing in Publication Data A catalogue record for this book is available from the British Library Copyright © 2019 by Morenike Oluwatoyin Folayan and contributors All rights for this book reserved. No part of this book may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without the prior permission of the copyright owner. ISBN (10): 1-5275-3691-2 ISBN (13): 978-1-5275-3691-3 TABLE OF CONTENTS Foreword .................................................................................................. viii Introduction ................................................................................................. 1 Dental Development: Anthropological Perspectives ................................. 31 Temitope A. Esan and Lynne A. Schepartz Belarus ....................................................................................................... 48 Natallia Shakavets, Alexander Yatzuk, Klavdia Gorbacheva and Nadezhda Chernyavskaya Bangladesh ............................................................................................... -

Insight Into the Role of Dental Pulp Stem Cells in Regenerative Therapy
biology Review Insight into the Role of Dental Pulp Stem Cells in Regenerative Therapy Shinichiro Yoshida 1,* , Atsushi Tomokiyo 1 , Daigaku Hasegawa 1, Sayuri Hamano 2,3, Hideki Sugii 1 and Hidefumi Maeda 1,3 1 Department of Endodontology, Kyushu University Hospital, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan; [email protected] (A.T.); [email protected] (D.H.); [email protected] (H.S.); [email protected] (H.M.) 2 OBT Research Center, Faculty of Dental Science, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan; [email protected] 3 Department of Endodontology and Operative Dentistry, Faculty of Dental Science, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan * Correspondence: [email protected]; Tel.: +81-92-642-6432 Received: 20 May 2020; Accepted: 5 July 2020; Published: 9 July 2020 Abstract: Mesenchymal stem cells (MSCs) have the capacity for self-renewal and multilineage differentiation potential, and are considered a promising cell population for cell-based therapy and tissue regeneration. MSCs are isolated from various organs including dental pulp, which originates from cranial neural crest-derived ectomesenchyme. Recently, dental pulp stem cells (DPSCs) and stem cells from human exfoliated deciduous teeth (SHEDs) have been isolated from dental pulp tissue of adult permanent teeth and deciduous teeth, respectively. Because of their MSC-like characteristics such as high growth capacity, multipotency, expression of MSC-related markers, and immunomodulatory effects, they are suggested to be an important cell source for tissue regeneration.