Evidence That Cryptomonad Chloroplasts Evolved from Photosynthetic Eukaryotic Endosymbionts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Apicoplast: a Review of the Derived Plastid of Apicomplexan Parasites
Curr. Issues Mol. Biol. 7: 57-80. Online journalThe Apicoplastat www.cimb.org 57 The Apicoplast: A Review of the Derived Plastid of Apicomplexan Parasites Ross F. Waller1 and Geoffrey I. McFadden2,* way to apicoplast discovery with studies of extra- chromosomal DNAs recovered from isopycnic density 1Botany, University of British Columbia, 3529-6270 gradient fractionation of total Plasmodium DNA. This University Boulevard, Vancouver, BC, V6T 1Z4, Canada group recovered two DNA forms; one a 6kb tandemly 2Plant Cell Biology Research Centre, Botany, University repeated element that was later identifed as the of Melbourne, 3010, Australia mitochondrial genome, and a second, 35kb circle that was supposed to represent the DNA circles previously observed by microscopists (Wilson et al., 1996b; Wilson Abstract and Williamson, 1997). This molecule was also thought The apicoplast is a plastid organelle, homologous to to be mitochondrial DNA, and early sequence data of chloroplasts of plants, that is found in apicomplexan eubacterial-like rRNA genes supported this organellar parasites such as the causative agents of Malaria conclusion. However, as the sequencing effort continued Plasmodium spp. It occurs throughout the Apicomplexa a new conclusion, that was originally embraced with and is an ancient feature of this group acquired by the some awkwardness (“Have malaria parasites three process of endosymbiosis. Like plant chloroplasts, genomes?”, Wilson et al., 1991), began to emerge. apicoplasts are semi-autonomous with their own genome Gradually, evermore convincing character traits of a and expression machinery. In addition, apicoplasts import plastid genome were uncovered, and strong parallels numerous proteins encoded by nuclear genes. These with plastid genomes from non-photosynthetic plants nuclear genes largely derive from the endosymbiont (Epifagus virginiana) and algae (Astasia longa) became through a process of intracellular gene relocation. -

Biochemical Characterization and Essentiality of Plasmodium
bioRxiv preprint doi: https://doi.org/10.1101/158956; this version posted July 3, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Biochemical characterization and essentiality of Plasmodium fumarate hydratase Vijay Jayaraman 1, Arpitha Suryavanshi 1, Pavithra Kalale1, Jyothirmayi Kunala1,2, Hemalatha Balaram 1# 1 Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR), Jakkur P.O., Bengaluru, Karnataka, 560064, INDIA. Tel: 91-80-22082812 Fax:91-80-22082766 2 Current affiliation: Molecular characterization/Analytical group, Biocon Research Ltd.-SEZ unit, Biocon Park, Bommasandra-Jigani link road, Bangalore, 560099, INDIA. #E-mail: [email protected] SUMMARY Plasmodium falciparum (Pf), the causative agent of malaria has an iron-sulfur cluster-containing class I fumarate hydratase (FH) that catalyzes the interconversion of fumarate to malate, a well-known reaction in the tricarboxylic acid cycle. In humans, the same reaction is catalyzed by class II FH that has no sequence or structural homology with the class I enzyme. Fumarate, generated in large quantities in the parasite as a byproduct of AMP synthesis is converted to malate by the action of FH, and subsequently used in the generation of the key metabolites oxaloacetate, aspartate and pyruvate. Here we report on the kinetic characterization of purified recombinant PfFH, functional complementation of fh deficiency in Escherichia. coli and mitochondrial localization in the parasite. The substrate analog, mercaptosuccinic acid was found to be a potent inhibitor of PfFH with a Ki value in the nanomolar range. -

Nanoplankton Protists from the Western Mediterranean Sea. II. Cryptomonads (Cryptophyceae = Cryptomonadea)*
sm69n1047 4/3/05 20:30 Página 47 SCI. MAR., 69 (1): 47-74 SCIENTIA MARINA 2005 Nanoplankton protists from the western Mediterranean Sea. II. Cryptomonads (Cryptophyceae = Cryptomonadea)* GIANFRANCO NOVARINO Department of Zoology, The Natural History Museum, Cromwell Road, London SW7 5BD, U.K. E-mail: [email protected] SUMMARY: This paper is an electron microscopical account of cryptomonad flagellates (Cryptophyceae = Cryptomon- adea) in the plankton of the western Mediterranean Sea. Bottle samples collected during the spring-summer of 1998 in the Sea of Alboran and Barcelona coastal waters contained a total of eleven photosynthetic species: Chroomonas (sensu aucto- rum) sp., Cryptochloris sp., 3 species of Hemiselmis, 3 species of Plagioselmis including Plagioselmis nordica stat. nov/sp. nov., Rhinomonas reticulata (Lucas) Novarino, Teleaulax acuta (Butcher) Hill, and Teleaulax amphioxeia (Conrad) Hill. Identification was based largely on cell surface features, as revealed by scanning electron microscopy (SEM). Cells were either dispersed in the water-column or associated with suspended particulate matter (SPM). Plagioselmis prolonga was the most common species both in the water-column and in association with SPM, suggesting that it might be a key primary pro- ducer of carbon. Taxonomic keys are given based on SEM. Key words: Cryptomonadea, cryptomonads, Cryptophyceae, flagellates, nanoplankton, taxonomy, ultrastructure. RESUMEN: PROTISTAS NANOPLANCTÓNICOS DEL MAR MEDITERRANEO NOROCCIDENTAL II. CRYPTOMONADALES (CRYPTOPHY- CEAE = CRYPTOMONADEA). – Este estudio describe a los flagelados cryptomonadales (Cryptophyceae = Cryptomonadea) planctónicos del Mar Mediterraneo Noroccidental mediante microscopia electrónica. La muestras recogidas en botellas durante la primavera-verano de 1998 en el Mar de Alboran y en aguas costeras de Barcelona, contenian un total de 11 espe- cies fotosintéticas: Chroomonas (sensu auctorum) sp., Cryptochloris sp., 3 especies de Hemiselmis, 3 especies de Plagio- selmis incluyendo Plagioselmis nordica stat. -

Sequestration, Performance, and Functional Control of Cryptophyte Plastids in the Ciliate Myrionecta Rubra (Ciliophora)1
J. Phycol. 42, 1235–1246 (2006) r 2006 by the Phycological Society of America DOI: 10.1111/j.1529-8817.2006.00275.x SEQUESTRATION, PERFORMANCE, AND FUNCTIONAL CONTROL OF CRYPTOPHYTE PLASTIDS IN THE CILIATE MYRIONECTA RUBRA (CILIOPHORA)1 Matthew D. Johnson2 Horn Point Laboratory, Center for Environmental Science, University of Maryland, Cambridge, Maryland 21613, USA Torstein Tengs National Veterinary Institute, Section of Food and Feed Microbiology, Ullevaalsveien 68, 0454 Oslo, Norway David Oldach Institute of Human Virology, School of Medicine, University of Maryland, Baltimore, Maryland, USA and Diane K. Stoecker Horn Point Laboratory, Center for Environmental Science, University of Maryland, Cambridge, Maryland 21613, USA Myrionecta rubra (Lohmann 1908, Jankowski Key index words: ciliate; Geminigera cryophila; 1976) is a photosynthetic ciliate with a global dis- mixotrophy; Myrionecta rubra; nucleomorph; or- tribution in neritic and estuarine habitats and has ganelle sequestration long been recognized to possess organelles of Abbreviations: CMC, chloroplast–mitochondria cryptophycean origin. Here we show, using nucleo- complex; HL, high light; LL, low light; LMWC, morph (Nm) small subunit rRNA gene sequence low-molecular-weight compound; MAA, micros- data, quantitative PCR, and pigment absorption scans, that an M. rubra culture has plastids identi- porine-like amino acids; ML, maximum likelihood; NGC, number of genomes per cell; PE, photosyn- cal to those of its cryptophyte prey, Geminigera thesis versus irradiance; TBR, tree bisection-recon- cf. cryophila (Taylor and Lee 1971, Hill 1991). Using quantitative PCR, we demonstrate that G. cf. struction cryophila plastids undergo division in growing M. rubra and are regulated by the ciliate. M. rubra maintained chl per cell and maximum cellu- Myrionecta rubra (5Mesodinium rubrum) (Lohmann cell lar photosynthetic rates (Pmax) that were 6–8 times 1908, Jankowski 1976) (Mesodiniidae, Litostomatea) that of G. -

Nuclear Genome Sequence of the Plastid-Lacking
Cenci et al. BMC Biology (2018) 16:137 https://doi.org/10.1186/s12915-018-0593-5 RESEARCH ARTICLE Open Access Nuclear genome sequence of the plastid- lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids Ugo Cenci1,2†, Shannon J. Sibbald1,2†, Bruce A. Curtis1,2, Ryoma Kamikawa3, Laura Eme1,2,11, Daniel Moog1,2,12, Bernard Henrissat4,5,6, Eric Maréchal7, Malika Chabi8, Christophe Djemiel8, Andrew J. Roger1,2,9, Eunsoo Kim10 and John M. Archibald1,2,9* Abstract Background: The evolution of photosynthesis has been a major driver in eukaryotic diversification. Eukaryotes have acquired plastids (chloroplasts) either directly via the engulfment and integration of a photosynthetic cyanobacterium (primary endosymbiosis) or indirectly by engulfing a photosynthetic eukaryote (secondary or tertiary endosymbiosis). The timing and frequency of secondary endosymbiosis during eukaryotic evolution is currently unclear but may be resolved in part by studying cryptomonads, a group of single-celled eukaryotes comprised of both photosynthetic and non-photosynthetic species. While cryptomonads such as Guillardia theta harbor a red algal-derived plastid of secondary endosymbiotic origin, members of the sister group Goniomonadea lack plastids. Here, we present the genome of Goniomonas avonlea—the first for any goniomonad—to address whether Goniomonadea are ancestrally non-photosynthetic or whether they lost a plastid secondarily. Results: We sequenced the nuclear and mitochondrial genomes of Goniomonas avonlea and carried out a comparative analysis of Go. avonlea, Gu. theta, and other cryptomonads. The Go. avonlea genome assembly is ~ 92 Mbp in size, with 33,470 predicted protein-coding genes. Interestingly, some metabolic pathways (e.g., fatty acid biosynthesis) predicted to occur in the plastid and periplastidal compartment of Gu. -

The Photosynthetic Endosymbiont in Cryptomonad Cells Produces Both Chloroplast and Cytoplasmic-Type Ribosomes
Journal of Cell Science 107, 649-657 (1994) 649 Printed in Great Britain © The Company of Biologists Limited 1994 JCS6601 The photosynthetic endosymbiont in cryptomonad cells produces both chloroplast and cytoplasmic-type ribosomes Geoffrey I. McFadden1,*, Paul R. Gilson1 and Susan E. Douglas2 1Plant Cell Biology Research Centre, School of Botany, University of Melbourne, Parkville, Victoria, 3052, Australia 2Institute of Marine Biosciences, National Research Council of Canada, 1411 Oxford St, Halifax, Nova Scotia B3H 3Z1, Canada *Author for correspondence SUMMARY Cryptomonad algae contain a photosynthetic, eukaryotic tion machinery. We also localized transcripts of the host endosymbiont. The endosymbiont is much reduced but nucleus rRNA gene. These transcripts were found in the retains a small nucleus. DNA from this endosymbiont nucleolus of the host nucleus, and throughout the host nucleus encodes rRNAs, and it is presumed that these cytoplasm, but never in the endosymbiont compartment. rRNAs are incorporated into ribosomes. Surrounding the Our rRNA localizations indicate that the cryptomonad cell endosymbiont nucleus is a small volume of cytoplasm produces two different of sets of cytoplasmic-type proposed to be the vestigial cytoplasm of the endosymbiont. ribosomes in two separate subcellular compartments. The If this compartment is indeed the endosymbiont’s results suggest that there is no exchange of rRNAs between cytoplasm, it would be expected to contain ribosomes with these compartments. We also used the probe specific for the components encoded by the endosymbiont nucleus. In this endosymbiont rRNA gene to identify chromosomes from paper, we used in situ hybridization to localize rRNAs the endosymbiont nucleus in pulsed field gel electrophore- encoded by the endosymbiont nucleus of the cryptomonad sis. -
Complete Nucleotide Sequence of the Chlorarachniophyte Nucleomorph: Nature’S Smallest Nucleus
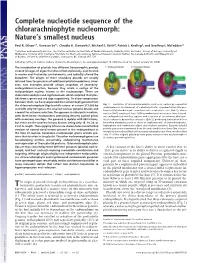
Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus Paul R. Gilson*†, Vanessa Su†‡, Claudio H. Slamovits§, Michael E. Reith¶, Patrick J. Keeling§, and Geoffrey I. McFadden‡ʈ *Infection and Immunity Division, The Walter and Eliza Hall Institute of Medical Research, Parkville 3050, Australia; ‡School of Botany, University of Melbourne, Victoria 3010, Australia; ¶Institute for Marine Biosciences, National Research Council, Halifax, NS, Canada B3H 3Z1; and §Department of Botany, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 Edited by Jeffrey D. Palmer, Indiana University, Bloomington, IN, and approved April 19, 2006 (received for review January 26, 2006) The introduction of plastids into different heterotrophic protists created lineages of algae that diversified explosively, proliferated in marine and freshwater environments, and radically altered the biosphere. The origins of these secondary plastids are usually inferred from the presence of additional plastid membranes. How- ever, two examples provide unique snapshots of secondary- endosymbiosis-in-action, because they retain a vestige of the endosymbiont nucleus known as the nucleomorph. These are chlorarachniophytes and cryptomonads, which acquired their plas- tids from a green and red alga respectively. To allow comparisons between them, we have sequenced the nucleomorph genome from the chlorarachniophyte Bigelowiella natans: at a mere 373,000 bp Fig. 1. Evolution of chlorarachniophytes such as B. natans by sequential endosymbioses. Enslavement of a photosynthetic, cyanobacterium-like pro- and with only 331 genes, the smallest nuclear genome known and karyote (Cb) introduces photosynthesis into a eukaryotic host (Euk 1), whose a model for extreme reduction. The genome is eukaryotic in nature, nucleus (Nu1) acquires at least 1,000 cyanobacterial genes over time. -

Red Algal Parasites: Models for a Life History Evolution That Leaves Photosynthesis Behind Again and Again
Prospects & Overviews Review essays Red algal parasites: Models for a life history evolution that leaves photosynthesis behind again and again Nicolas A. Blouinà and Christopher E. Lane Many of the most virulent and problematic eukaryotic Introduction pathogens have evolved from photosynthetic ancestors, such as apicomplexans, which are responsible for a Parasitology is one of the oldest fields of medical research and continues to be an essential area of study on organisms wide range of diseases including malaria and toxoplas- that kill millions annually, either directly or through mosis. The primary barrier to understanding the early agricultural loss. In the early genomics era, parasites were stages of evolution of these parasites has been the diffi- some of the initial eukaryotes to have their genomes culty in finding parasites with closely related free-living sequenced. The combination of medical interest and small lineages with which to make comparisons. Parasites genome size (due to genome compaction [1]) has resulted found throughout the florideophyte red algal lineage, in a relatively large number of sequenced genomes from these taxa. The range of relationships that exist between however, provide a unique and powerful model to inves- parasites and comparative free-living taxa, however, compli- tigate the genetic origins of a parasitic lifestyle. This is cates understanding the evolution of eukaryotic parasitism. because they share a recent common ancestor with an In some cases (such as apicomplexans, which cause extant free-living red algal species and parasitism has malaria, cryptosporidiosis and toxoplasmosis, among other independently arisen over 100 times within this group. diseases) entire lineages appear to have a common parasitic ancestor [2]. -

Systema Naturae. the Classification of Living Organisms
Systema Naturae. The classification of living organisms. c Alexey B. Shipunov v. 5.601 (June 26, 2007) Preface Most of researches agree that kingdom-level classification of living things needs the special rules and principles. Two approaches are possible: (a) tree- based, Hennigian approach will look for main dichotomies inside so-called “Tree of Life”; and (b) space-based, Linnaean approach will look for the key differences inside “Natural System” multidimensional “cloud”. Despite of clear advantages of tree-like approach (easy to develop rules and algorithms; trees are self-explaining), in many cases the space-based approach is still prefer- able, because it let us to summarize any kinds of taxonomically related da- ta and to compare different classifications quite easily. This approach also lead us to four-kingdom classification, but with different groups: Monera, Protista, Vegetabilia and Animalia, which represent different steps of in- creased complexity of living things, from simple prokaryotic cell to compound Nature Precedings : doi:10.1038/npre.2007.241.2 Posted 16 Aug 2007 eukaryotic cell and further to tissue/organ cell systems. The classification Only recent taxa. Viruses are not included. Abbreviations: incertae sedis (i.s.); pro parte (p.p.); sensu lato (s.l.); sedis mutabilis (sed.m.); sedis possi- bilis (sed.poss.); sensu stricto (s.str.); status mutabilis (stat.m.); quotes for “environmental” groups; asterisk for paraphyletic* taxa. 1 Regnum Monera Superphylum Archebacteria Phylum 1. Archebacteria Classis 1(1). Euryarcheota 1 2(2). Nanoarchaeota 3(3). Crenarchaeota 2 Superphylum Bacteria 3 Phylum 2. Firmicutes 4 Classis 1(4). Thermotogae sed.m. 2(5). -

Early Diverging Lineages Within Cryptomycota and Chytridiomycota Dominate the Fungal Communities in Ice-Covered Lakes of the Mcmurdo Dry Valleys, Antarctica
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/320986652 Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities in ice-covered lakes of the McMurdo Dry Valleys, Antarctica Article in Scientific Reports · November 2017 DOI: 10.1038/s41598-017-15598-w CITATIONS READS 2 144 6 authors, including: Keilor Rojas- Jimenez Christian Wurzbacher University of Costa Rica Technische Universität München 28 PUBLICATIONS 289 CITATIONS 59 PUBLICATIONS 398 CITATIONS SEE PROFILE SEE PROFILE Elizabeth Bourne Amy Chiuchiolo Leibniz-Institute of Freshwater Ecology and Inland Fisheries Montana State University 9 PUBLICATIONS 450 CITATIONS 11 PUBLICATIONS 322 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: MANTEL View project HGT in aquatic ecosystems View project All content following this page was uploaded by Keilor Rojas-Jimenez on 10 November 2017. The user has requested enhancement of the downloaded file. www.nature.com/scientificreports OPEN Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities Received: 25 August 2017 Accepted: 30 October 2017 in ice-covered lakes of the McMurdo Published: xx xx xxxx Dry Valleys, Antarctica Keilor Rojas-Jimenez 1,2, Christian Wurzbacher1,3, Elizabeth Charlotte Bourne3,4, Amy Chiuchiolo5, John C. Priscu5 & Hans-Peter Grossart 1,6 Antarctic ice-covered lakes are exceptional sites for studying the ecology of aquatic fungi under conditions of minimal human disturbance. In this study, we explored the diversity and community composition of fungi in fve permanently covered lake basins located in the Taylor and Miers Valleys of Antarctica. -

Nuclear Genome Sequence of the Plastid
Nuclear genome sequence of the plastid-lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids Ugo Cenci, Shannon Sibbald, Bruce Curtis, Ryoma Kamikawa, Laura Eme, Daniel Moog, Bernard Henrissat, Eric Marechal, Malika Chabi, Christophe Djemiel, et al. To cite this version: Ugo Cenci, Shannon Sibbald, Bruce Curtis, Ryoma Kamikawa, Laura Eme, et al.. Nuclear genome sequence of the plastid-lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids. BMC Biology, BioMed Central, 2018, 16 (1), pp.137. 10.1186/s12915-018- 0593-5. hal-02046523 HAL Id: hal-02046523 https://hal.archives-ouvertes.fr/hal-02046523 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Cenci et al. BMC Biology (2018) 16:137 https://doi.org/10.1186/s12915-018-0593-5 RESEARCH ARTICLE Open Access Nuclear genome sequence of the plastid- lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids Ugo Cenci1,2†, Shannon J. Sibbald1,2†, Bruce A. Curtis1,2, Ryoma Kamikawa3, Laura Eme1,2,11, Daniel Moog1,2,12, Bernard Henrissat4,5,6, Eric Maréchal7, Malika Chabi8, Christophe Djemiel8, Andrew J. -

Phylogenetic Profiles of All Membrane Transport Proteins of the Malaria Parasite Highlight New Drug Targets
Research Article www.microbialcell.com Phylogenetic profiles of all membrane transport proteins of the malaria parasite highlight new drug targets January Weiner 3rd 1 and Taco W.A. Kooij 2, * 1 Department of Immunology, Max Planck Institute for Infection Biology, Berlin, Germany. 2 Department of Medical Microbiology & Centre for Molecular and Biomolecular Informatics, Radboud Institute for Molecular Life Sciences, Radboud University Medical Centre, Nijmegen, The Netherlands. * Corresponding Author: Taco W.A. Kooij, Department of Medical Microbiology & Centre for Molecular and Biomolecular Informatics, Radboud Institute for Molecular Life Sciences, Radboud University Medical Centre; P.O. Box 9101, 6500 HB Nijmegen, The Netherlands; Tel: +31 24 36 10113; Fax: +31 24 36 19395; E-mail: [email protected] ABSTRACT In order to combat the on-going malaria epidemic, discovery of doi: 10.15698/mic2016.10.534 new drug targets remains vital. Proteins that are essential to survival and spe- Received originally: 01 .04.2016 ; cific to malaria parasites are key candidates. To survive within host cells, the in revised form: 30.07.2016, Accepted 01 .08.2016, parasites need to acquire nutrients and dispose of waste products across mul- Published 30.08.2016. tiple membranes. Additionally, like all eukaryotes, they must redistribute ions and organic molecules between their various internal membrane bound com- partments. Membrane transport proteins mediate all of these processes and Keywords : drug target, experimental are considered important mediators of drug resistance as well as drug targets genetics, malaria parasite, membrane in their own right. Recently, using advanced experimental genetic approaches transport protein, orthology, phylogeny, Plasmodium. and streamlined life cycle profiling, we generated a large collection of Plas- modium berghei gene deletion mutants and assigned essential gene func- tions, highlighting potential targets for prophylactic, therapeutic, and trans- mission-blocking anti-malarial drugs.