Complete Mitochondrial Genome of Callogorgia Cf. Gracilis (Octocorallia: Calcaxonia: Primnoidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Time-Calibrated Molecular Phylogeny of the Precious Corals: Reconciling Discrepancies in the Taxonomic Classification and Insights Into Their Evolutionary History
A time-calibrated molecular phylogeny of the precious corals: reconciling discrepancies in the taxonomic classification and insights into their evolutionary history The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Ardila, Néstor E, Gonzalo Giribet, and Juan A Sánchez. 2012. A time- calibrated molecular phylogeny of the precious corals: reconciling discrepancies in the taxonomic classification and insights into their evolutionary history. BMC Evolutionary Biology 12: 246. Published Version doi:10.1186/1471-2148-12-246 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:11802886 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA A time-calibrated molecular phylogeny of the precious corals: reconciling discrepancies in the taxonomic classification and insights into their evolutionary history Ardila et al. Ardila et al. BMC Evolutionary Biology 2012, 12:246 http://www.biomedcentral.com/1471-2148/12/246 Ardila et al. BMC Evolutionary Biology 2012, 12:246 http://www.biomedcentral.com/1471-2148/12/246 RESEARCH ARTICLE Open Access A time-calibrated molecular phylogeny of the precious corals: reconciling discrepancies in the taxonomic classification and insights into their evolutionary history Néstor E Ardila1,3, Gonzalo Giribet2 and Juan A Sánchez1* Abstract Background: Seamount-associated faunas are often considered highly endemic but isolation and diversification processes leading to such endemism have been poorly documented at those depths. -

Habitat-Forming Deep-Sea Corals in the Northeast Pacific Ocean
Habitat-forming deep-sea corals in the Northeast Pacific Ocean Peter Etnoyer1, Lance E. Morgan2 1 Aquanautix Consulting, 3777 Griffith View Drive, Los Angeles, CA 90039, USA ([email protected]) 2 Marine Conservation Biology Institute, 4878 Warm Springs Rd., Glen Ellen, CA 95442, USA Abstract. We define habitat-forming deep-sea corals as those families of octocorals, hexacorals, and stylasterids with species that live deeper than 200 m, with a majority of species exhibiting complex branching morphology and a sufficient size to provide substrata or refugia to associated species. We present 2,649 records (name, geoposition, depth, and data quality) from eleven institutions on eight habitat- forming deep-sea coral families, including octocorals in the families Coralliidae, Isididae, Paragorgiidae and Primnoidae, hexacorals in the families Antipathidae, Oculinidae and Caryophylliidae, and stylasterids in the family Stylasteridae. The data are ranked according to record quality. We compare family range and distribution as predicted by historical records to the family extent as informed by recent collections aboard the National Oceanic of Atmospheric Administration (NOAA) Office of Ocean Exploration 2002 Gulf of Alaska Seamount Expedition (GOASEX). We present a map of one of these families, the Primnoidae. We find that these habitat-forming families are widespread throughout the Northeast Pacific, save Caryophylliidae (Lophelia sp.) and Oculinidae (Madrepora sp.), which are limited in occurrence. Most coral records fall on the continental shelves, in Alaska, or Hawaii, likely reflecting research effort. The vertical range of these families, based on large samples (N >200), is impressive. Four families have maximum-recorded depths deeper than 1500 m, and minimum depths shallower than 40 m. -
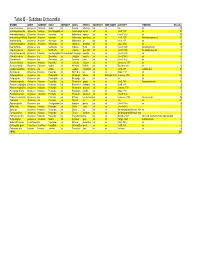
Table B – Subclass Octocorallia
Table B – Subclass Octocorallia BINOMEN ORDER SUBORDER FAMILY SUBFAMILY GENUS SPECIES SUBSPECIES COMN_NAMES AUTHORITY SYNONYMS #Records Acanella arbuscula Alcyonacea Calcaxonia Isididae n/a Acanella arbuscula n/a n/a n/a n/a 59 Acanthogorgia armata Alcyonacea Holaxonia Acanthogorgiidae n/a Acanthogorgia armata n/a n/a Verrill, 1878 n/a 95 Anthomastus agassizii Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus agassizii n/a n/a (Verrill, 1922) n/a 35 Anthomastus grandiflorus Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus grandiflorus n/a n/a Verrill, 1878 Anthomastus purpureus 37 Anthomastus sp. Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus sp. n/a n/a Verrill, 1878 n/a 1 Anthothela grandiflora Alcyonacea Scleraxonia Anthothelidae n/a Anthothela grandiflora n/a n/a (Sars, 1856) n/a 24 Capnella florida Alcyonacea n/a Nephtheidae n/a Capnella florida n/a n/a (Verrill, 1869) Eunephthya florida 44 Capnella glomerata Alcyonacea n/a Nephtheidae n/a Capnella glomerata n/a n/a (Verrill, 1869) Eunephthya glomerata 4 Chrysogorgia agassizii Alcyonacea Holaxonia Acanthogorgiidae Chrysogorgiidae Chrysogorgia agassizii n/a n/a (Verrill, 1883) n/a 2 Clavularia modesta Alcyonacea n/a Clavulariidae n/a Clavularia modesta n/a n/a (Verrill, 1987) n/a 6 Clavularia rudis Alcyonacea n/a Clavulariidae n/a Clavularia rudis n/a n/a (Verrill, 1922) n/a 1 Gersemia fruticosa Alcyonacea Alcyoniina Alcyoniidae n/a Gersemia fruticosa n/a n/a Marenzeller, 1877 n/a 3 Keratoisis flexibilis Alcyonacea Calcaxonia Isididae n/a Keratoisis flexibilis n/a n/a Pourtales, 1868 n/a 1 Lepidisis caryophyllia Alcyonacea n/a Isididae n/a Lepidisis caryophyllia n/a n/a Verrill, 1883 Lepidisis vitrea 13 Muriceides sp. -

Tree Coral, Primnoa Pacifica
Sexual Reproduction and Seasonality of the Alaskan Red Tree Coral, Primnoa pacifica Rhian G. Waller1*, Robert P. Stone2, Julia Johnstone3, Jennifer Mondragon4 1 University of Maine, School of Marine Sciences, Darling Marine Center, Walpole, Maine, United States of America, 2 Alaska Fisheries Science Center, National Marine Fisheries Service, National Oceanic and Atmospheric Administration, Juneau, Alaska, United States of America, 3 University of Maine, Darling Marine Center, Walpole, Maine, United States of America, 4 Alaska Regional Office, National Marine Fisheries Service, National Oceanic and Atmospheric Administration, Juneau, Alaska, United States of America Abstract The red tree coral Primnoa pacifica is an important habitat forming octocoral in North Pacific waters. Given the prominence of this species in shelf and upper slope areas of the Gulf of Alaska where fishing disturbance can be high, it may be able to sustain healthy populations through adaptive reproductive processes. This study was designed to test this hypothesis, examining reproductive mode, seasonality and fecundity in both undamaged and simulated damaged colonies over the course of 16 months using a deepwater-emerged population in Tracy Arm Fjord. Females within the population developed asynchronously, though males showed trends of synchronicity, with production of immature spermatocysts heightened in December/January and maturation of gametes in the fall months. Periodicity of individuals varied from a single year reproductive event to some individuals taking more than the 16 months sampled to produce viable gametes. Multiple stages of gametes occurred in polyps of the same colony during most sampling periods. Mean oocyte size ranged from 50 to 200 mm in any season, and maximum oocyte size (802 mm) suggests a lecithotrophic larva. -

Deep-Sea Origin and In-Situ Diversification of Chrysogorgiid Octocorals
Deep-Sea Origin and In-Situ Diversification of Chrysogorgiid Octocorals Eric Pante1*¤, Scott C. France1, Arnaud Couloux2, Corinne Cruaud2, Catherine S. McFadden3, Sarah Samadi4, Les Watling5,6 1 Department of Biology, University of Louisiana at Lafayette, Lafayette, Louisiana, United States of America, 2 GENOSCOPE, Centre National de Se´quenc¸age, Evry, France, 3 Department of Biology, Harvey Mudd College, Claremont, California, United States of America, 4 De´partement Syste´matique et Evolution, UMR 7138 UPMC-IRD-MNHN- CNRS (UR IRD 148), Muse´um national d’Histoire naturelle, Paris, France, 5 Department of Biology, University of Hawaii at Manoa, Honolulu, Hawaii, United States of America, 6 Darling Marine Center, University of Maine, Walpole, Maine, United States of America Abstract The diversity, ubiquity and prevalence in deep waters of the octocoral family Chrysogorgiidae Verrill, 1883 make it noteworthy as a model system to study radiation and diversification in the deep sea. Here we provide the first comprehensive phylogenetic analysis of the Chrysogorgiidae, and compare phylogeny and depth distribution. Phylogenetic relationships among 10 of 14 currently-described Chrysogorgiidae genera were inferred based on mitochondrial (mtMutS, cox1) and nuclear (18S) markers. Bathymetric distribution was estimated from multiple sources, including museum records, a literature review, and our own sampling records (985 stations, 2345 specimens). Genetic analyses suggest that the Chrysogorgiidae as currently described is a polyphyletic family. Shallow-water genera, and two of eight deep-water genera, appear more closely related to other octocoral families than to the remainder of the monophyletic, deep-water chrysogorgiid genera. Monophyletic chrysogorgiids are composed of strictly (Iridogorgia Verrill, 1883, Metallogorgia Versluys, 1902, Radicipes Stearns, 1883, Pseudochrysogorgia Pante & France, 2010) and predominantly (Chrysogorgia Duchassaing & Michelotti, 1864) deep-sea genera that diversified in situ. -

Guide to the Identification of Precious and Semi-Precious Corals in Commercial Trade
'l'llA FFIC YvALE ,.._,..---...- guide to the identification of precious and semi-precious corals in commercial trade Ernest W.T. Cooper, Susan J. Torntore, Angela S.M. Leung, Tanya Shadbolt and Carolyn Dawe September 2011 © 2011 World Wildlife Fund and TRAFFIC. All rights reserved. ISBN 978-0-9693730-3-2 Reproduction and distribution for resale by any means photographic or mechanical, including photocopying, recording, taping or information storage and retrieval systems of any parts of this book, illustrations or texts is prohibited without prior written consent from World Wildlife Fund (WWF). Reproduction for CITES enforcement or educational and other non-commercial purposes by CITES Authorities and the CITES Secretariat is authorized without prior written permission, provided the source is fully acknowledged. Any reproduction, in full or in part, of this publication must credit WWF and TRAFFIC North America. The views of the authors expressed in this publication do not necessarily reflect those of the TRAFFIC network, WWF, or the International Union for Conservation of Nature (IUCN). The designation of geographical entities in this publication and the presentation of the material do not imply the expression of any opinion whatsoever on the part of WWF, TRAFFIC, or IUCN concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The TRAFFIC symbol copyright and Registered Trademark ownership are held by WWF. TRAFFIC is a joint program of WWF and IUCN. Suggested citation: Cooper, E.W.T., Torntore, S.J., Leung, A.S.M, Shadbolt, T. and Dawe, C. -

Anthozoa: Octocorallia) from the Clarion-Clipperton Fracture Zone, Equatorial Northeastern Pacific
Mar Biodiv DOI 10.1007/s12526-015-0340-x ORIGINAL PAPER New abyssal Primnoidae (Anthozoa: Octocorallia) from the Clarion-Clipperton Fracture Zone, equatorial northeastern Pacific Stephen D. Cairns1 Received: 28 January 2015 /Revised: 20 March 2015 /Accepted: 23 March 2015 # Senckenberg Gesellschaft für Naturforschung and Springer-Verlag Berlin Heidelberg 2015 Abstract Three new species, including a new genus, nodules that contain iron, manganese, copper, nickel, cobalt, Abyssoprimnoa, are described from abyssal depths from the zinc, silver, and other metals (Bluhm 1994), and thus have easternmost Clarion-Clipperton Fracture Zone in the equato- been of interest to deep-sea mining companies for several rial northeastern Pacific. This prompted the listing of all 39 decades since the region was discovered in the 1950s. octocorallian taxa collected deeper than 3000 m, which con- Currently, at least 12 claim sites have been awarded to various stitutes only about 1.2 % of the octocoral species. To place this mining companies, each associated with one or more coun- in perspective, the depth records for other benthic cnidarian tries. The hard substrate afforded by the nodules allows the orders are compared. settlement of various benthic invertebrates that require a hard surface for attachment and subsequent anchoring, one of these groups being the octocorals. Because of the extreme depth and Keywords Primnoidae . Clarion-clipperton fracture zone . logistic difficulty of collecting organisms from such depths, New species . New genus . Octocorallia -

Deep‐Sea Coral Taxa in the U.S. Gulf of Mexico: Depth and Geographical Distribution
Deep‐Sea Coral Taxa in the U.S. Gulf of Mexico: Depth and Geographical Distribution by Peter J. Etnoyer1 and Stephen D. Cairns2 1. NOAA Center for Coastal Monitoring and Assessment, National Centers for Coastal Ocean Science, Charleston, SC 2. National Museum of Natural History, Smithsonian Institution, Washington, DC This annex to the U.S. Gulf of Mexico chapter in “The State of Deep‐Sea Coral Ecosystems of the United States” provides a list of deep‐sea coral taxa in the Phylum Cnidaria, Classes Anthozoa and Hydrozoa, known to occur in the waters of the Gulf of Mexico (Figure 1). Deep‐sea corals are defined as azooxanthellate, heterotrophic coral species occurring in waters 50 m deep or more. Details are provided on the vertical and geographic extent of each species (Table 1). This list is adapted from species lists presented in ʺBiodiversity of the Gulf of Mexicoʺ (Felder & Camp 2009), which inventoried species found throughout the entire Gulf of Mexico including areas outside U.S. waters. Taxonomic names are generally those currently accepted in the World Register of Marine Species (WoRMS), and are arranged by order, and alphabetically within order by suborder (if applicable), family, genus, and species. Data sources (references) listed are those principally used to establish geographic and depth distribution. Only those species found within the U.S. Gulf of Mexico Exclusive Economic Zone are presented here. Information from recent studies that have expanded the known range of species into the U.S. Gulf of Mexico have been included. The total number of species of deep‐sea corals documented for the U.S. -

Phylogenetic Relationships Within Chrysogorgia (Alcyonacea
Phylogenetic Relationships Within Chrysogorgia (Alcyonacea: Octocorallia), a Morphologically Diverse Genus of Octocoral, Revealed Using a Target Enrichment Approach Candice Untiedt, Andrea Quattrini, Catherine Mcfadden, Phil Alderslade, Eric Pante, Christopher Burridge To cite this version: Candice Untiedt, Andrea Quattrini, Catherine Mcfadden, Phil Alderslade, Eric Pante, et al.. Phy- logenetic Relationships Within Chrysogorgia (Alcyonacea: Octocorallia), a Morphologically Diverse Genus of Octocoral, Revealed Using a Target Enrichment Approach. Frontiers in Marine Science, Frontiers Media, 2021, 7, 10.3389/fmars.2020.599984. hal-03154113 HAL Id: hal-03154113 https://hal.sorbonne-universite.fr/hal-03154113 Submitted on 27 Feb 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. fmars-07-599984 January 11, 2021 Time: 10:55 # 1 ORIGINAL RESEARCH published: 12 January 2021 doi: 10.3389/fmars.2020.599984 Phylogenetic Relationships Within Chrysogorgia (Alcyonacea: Octocorallia), a Morphologically Diverse Genus of Octocoral, Revealed Using a Target Enrichment -
![Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill](https://docslib.b-cdn.net/cover/9917/genetic-divergence-and-polyphyly-in-the-octocoral-genus-swiftia-cnidaria-octocorallia-including-a-species-impacted-by-the-dwh-oil-spill-739917.webp)
Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill
diversity Article Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill Janessy Frometa 1,2,* , Peter J. Etnoyer 2, Andrea M. Quattrini 3, Santiago Herrera 4 and Thomas W. Greig 2 1 CSS Dynamac, Inc., 10301 Democracy Lane, Suite 300, Fairfax, VA 22030, USA 2 Hollings Marine Laboratory, NOAA National Centers for Coastal Ocean Sciences, National Ocean Service, National Oceanic and Atmospheric Administration, 331 Fort Johnson Rd, Charleston, SC 29412, USA; [email protected] (P.J.E.); [email protected] (T.W.G.) 3 Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, 10th and Constitution Ave NW, Washington, DC 20560, USA; [email protected] 4 Department of Biological Sciences, Lehigh University, 111 Research Dr, Bethlehem, PA 18015, USA; [email protected] * Correspondence: [email protected] Abstract: Mesophotic coral ecosystems (MCEs) are recognized around the world as diverse and ecologically important habitats. In the northern Gulf of Mexico (GoMx), MCEs are rocky reefs with abundant black corals and octocorals, including the species Swiftia exserta. Surveys following the Deepwater Horizon (DWH) oil spill in 2010 revealed significant injury to these and other species, the restoration of which requires an in-depth understanding of the biology, ecology, and genetic diversity of each species. To support a larger population connectivity study of impacted octocorals in the Citation: Frometa, J.; Etnoyer, P.J.; GoMx, this study combined sequences of mtMutS and nuclear 28S rDNA to confirm the identity Quattrini, A.M.; Herrera, S.; Greig, Swiftia T.W. -

A Possible Method for Improving the Conservation Status of a Ellisella
Research Article Mediterranean Marine Science Indexed in WoS (Web of Science, ISI Thomson) and SCOPUS The journal is available on line at http://www.medit-mar-sc.net http://dx.doi.org/10.12681/mms.2076 Pruning treatment: A possible method for improving the conservation status of a Ellisella paraplexauroides Stiasny, 1936 (Anthozoa, Alcyonacea) population in the Chafarinas Islands? LUIS SÁNCHEZ -TOCINO1, ANTONIO DE LA LINDE RUBIO2, M SOL LIZANA ROSAS1, TEODORO PÉREZ GUERRA1 and JOSE MANUEL TIERNO DE FIGUEROA1 1 Departamento de Zoología. Facultad de Ciencias. Universidad de Granada. Campus Fuentenueva s/n, 18071, Granada, Spain 2 Urbanización los Delfines, Pl 4, 2º, 11207, Algeciras, Cádiz, Spain Corresponding author: [email protected] Handling Editor: Emma Cebrian Received: 7 December 2017; Accepted: 11 July 2017; Published on line: 11 December 2017 Abstract In the present paper, the results of a four-year conservation study on the giant gorgonian Ellisella paraplexauroides in the Chafarinas Islands are reported. This species, currently protected in Spain, is present as isolated colonies or as a low number of them, except in the Chafarinas Islands, where higher densities can be found below a depth of 20 m. Nevertheless, as revealed by a previous study, a great number of colonies are partially covered by epibionts or totally dead. As a first objective of the present work, seven transects were performed in 2013 and 2014 to evaluate the percentage of colonies affected in areas that had not been previously sampled. Approximately between 35% and 95% of the colonies had different degrees of epibiosis or were dead. In 2015, ten transects were performed to specifically locate young colonies smaller than 15 cm, which could be indicative of popula- tion regeneration by sexual reproduction. -

Primnoidae (Octocorallia: Calcaxonia) from the Emperor Seamounts, with Notes on Callogorgia Elegans (Gray, 1870)
Primnoidae (Octocorallia: Calcaxonia) from the Emperor Seamounts, with Notes on Callogorgia elegans (Gray, 1870) Stephen D. Cairns, Robert P. Stone, Hye-Won Moon, Jong Hee Lee Pacific Science, Volume 72, Number 1, January 2018, pp. 125-142 (Article) Published by University of Hawai'i Press For additional information about this article https://muse.jhu.edu/article/683173 [ This content has been declared free to read by the pubisher during the COVID-19 pandemic. ] Primnoidae (Octocorallia: Calcaxonia) from the Emperor Seamounts, with Notes on Callogorgia elegans (Gray, 1870)1 Stephen D. Cairns,2,6 Robert P. Stone,3 Hye-Won Moon,4 and Jong Hee Lee 5 Abstract: Six primnoid species are reported from depths of 280 – 480 m from the southern Emperor Seamounts, including two new species (Callogorgia imperialis and Thouarella taylorae). Only the new species are fully described and illustrated. Also, Callogorgia elegans, which has a confused taxonomic history, is discussed and illustrated. Not unexpectedly, the Emperor Seamount primnoids have a strong affinity with the fauna of the Hawaiian Islands, an affinity that is expected to increase as more collecting is done in the region. The United Nations General Assembly nations around the world are developing pro- approved Resolution 61/105 in December tocol and policy on fishing encounters with 2006 ( United Nations General Assembly 2007) the sensitive biota (Durán Muñoz et al. 2012). that calls on States to directly, or through Here we report on collections made on fish- Regional Fisheries Management Organiza- ing vessels in the Emperor Seamounts, North tions, apply a precautionary ecosystem ap- Pacific Ocean, as part of a joint project be- proach to sustainably manage fish stocks and tween the United States and the Republic of protect vulnerable marine ecosystems ( VMEs) Korea.