A Paley, Front Matter, FINAL
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

William D. Mcelroy Papers
http://oac.cdlib.org/findaid/ark:/13030/kt6489r0v5 No online items William D. McElroy Papers Special Collections & Archives, UC San Diego Special Collections & Archives, UC San Diego Copyright 2005 9500 Gilman Drive La Jolla 92093-0175 [email protected] URL: http://libraries.ucsd.edu/collections/sca/index.html William D. McElroy Papers MSS 0483 1 Descriptive Summary Languages: English Contributing Institution: Special Collections & Archives, UC San Diego 9500 Gilman Drive La Jolla 92093-0175 Title: William D. McElroy Papers Identifier/Call Number: MSS 0483 Physical Description: 10.4 Linear feet(19 archives boxes, 11 oversize folders, 6 art bin items) Date (inclusive): 1944-1999 Abstract: Papers of William David McElroy (1917-1999), professor of biochemistry, the fourth chancellor (1972-1980) of the University of California, San Diego; and former director (1969-1971) of the National Science Foundation. Scope and Content of Collection Papers of William David McElroy (1917-1999), professor of biochemistry, the fourth chancellor (1972-1980) of the University of California, San Diego; and former director (1969-1971) of the National Science Foundation. McElroy's significant contributions to biology include isolating and crystallizing the compounds that enable firefly luminescence and for his subsequent research into bacterial bioluminescence. He also wrote, spoke, and worked on problems in areas of environment, pollution, food production, science education, and international science. The papers largely document McElroy's scientific research and include correspondence with the scientific community, various biographical materials including awards and photographs, his trip to China in 1979, writings and reprints related to biochemical and scientific investigation, research materials on bioluminescence, teaching materials, and speeches given both as chancellor and as director of the National Science Foundation. -

Safety Data Sheet
G-Biosciences, St Louis, MO, USA | 1-800-628-7730 | 1-314-991-6034 | [email protected] A Geno Technology, Inc. (USA) brand name Safety Data Sheet Cat. # RC-227 D-Luciferin Firefly, FREE ACID Size: 0.25g think proteins! think G-Biosciences! www.GBiosciences.com D-Luciferin Firefly, potassium salt Safety Data Sheet according to Federal Register / Vol. 77, No. 58 / Monday, March 26, 2012 / Rules and Regulations Date of issue: 05/06/2013 Revision date: 05/11/2017 Version: 7.1 SECTION 1: Identification 1.1. Identification Product form : Substance Trade name : D-Luciferin Firefly, potassium salt CAS-No. : 2591-17-5 Product code : 006A Formula : C11H8N2O3S2 Synonyms : (4S)-4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-4-thiazolecarboxylic acid / (S)-2-(6-hydroxy-2- benzothiazolyl)-2-thiazoline-4-carboxylic acid / (S)-4,5-dihydro-2-(6-hydroxybenzothiazol-2- yl)thiazole-4-carboxylic acid / 2-(6-hydroxy-2-benzothiazolyl)-2-thiazoline-4-carcoxylic acid, (S)- / 2-(6-hydroxybenzothiazol-2-yl)-2-thiazoline-4-carboxylic acid / 4,5-dihydro-2-(6-hydroxy-2- benzorhiazolyl)-4-thiazolecarboxylic acid, (4S)- / 4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-4- thiazolecarboxylic acid / 4-Thiazolecarboxylic acid, 4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-, (S)- / D-(-)-luciferin / D-luciferin / firefly luciferin / liciferin, D-(-)- / luciferin / luciferin, D- Other means of identification : D-Luciferin Firefly, free acid 4,5-Dihydro-2-(6-hydroxy-2-benzothiazolyl)-4-thiazolecarboxylic acid, ST50405784, Luciferin, CHEBI:17165 BIG No : 48631 1.2. Recommended use and restrictions on use Use of the substance/mixture : Luciferin is a common bioluminescent reporter used for in-vivo imaging of the expression of the luc marker gene . -

Bioluminescence Is Produced by a Firefly-Like Luciferase but an Entirely
www.nature.com/scientificreports OPEN New Zealand glowworm (Arachnocampa luminosa) bioluminescence is produced by a Received: 8 November 2017 Accepted: 1 February 2018 frefy-like luciferase but an entirely Published: xx xx xxxx new luciferin Oliver C. Watkins1,2, Miriam L. Sharpe 1, Nigel B. Perry 2 & Kurt L. Krause 1 The New Zealand glowworm, Arachnocampa luminosa, is well-known for displays of blue-green bioluminescence, but details of its bioluminescent chemistry have been elusive. The glowworm is evolutionarily distant from other bioluminescent creatures studied in detail, including the frefy. We have isolated and characterised the molecular components of the glowworm luciferase-luciferin system using chromatography, mass spectrometry and 1H NMR spectroscopy. The purifed luciferase enzyme is in the same protein family as frefy luciferase (31% sequence identity). However, the luciferin substrate of this enzyme is produced from xanthurenic acid and tyrosine, and is entirely diferent to that of the frefy and known luciferins of other glowing creatures. A candidate luciferin structure is proposed, which needs to be confrmed by chemical synthesis and bioluminescence assays. These fndings show that luciferases can evolve independently from the same family of enzymes to produce light using structurally diferent luciferins. Glowworms are found in New Zealand and Australia, and are a major tourist attraction at sites located across both countries. In contrast to luminescent beetles such as the frefy (Coleoptera), whose bioluminescence has been well characterised (reviewed by ref.1), the molecular details of glowworm bioluminescence have remained elusive. Tese glowworms are the larvae of fungus gnats of the genus Arachnocampa, with eight species endemic to Australia and a single species found only in New Zealand2. -
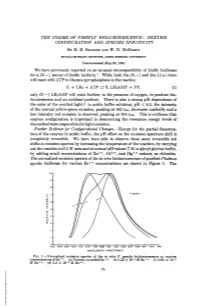
The Colors of Firefly Bioluminescence: Enzyme Configuration and Species Specificity by H
THE COLORS OF FIREFLY BIOLUMINESCENCE: ENZYME CONFIGURATION AND SPECIES SPECIFICITY BY H. H. SELIGER AND W. D. MCELROY MCCOLLUM-PRATT INSTITUTE, JOHNS HOPKINS UNIVERSITY Communicated May 25, 1964 We have previously reported on an unusual stereospecificity of firefly luciferase for a D(-) isomer of firefly luciferin.' While both the D(-) and the L(+) form will react with ATP to liberate pyrophosphate in the reaction E + LH2 + ATP =- E. LH2AMP + PP, (1) only D(-) LH2AMP will react further, in the presence of oxygen, to produce bio- luminescence and an oxidized product. There is also a strong pH dependence of the color of the emitted light;2 in acidic buffer solutions, pH < 6.5, the intensity of the normal yellow-green emission, peaking at 562 ml,, decreases markedly and a low intensity red emission is observed, peaking at 616 miu. This is evidence that enzyme configuration is important in determining the resonance energy levels of the excited state responsible for light emission. Further Evidence for Configurational Changes.-Except for the partial denatura- tion of the enzyme in acidic buffer, the pH effect on the emission spectrum shift is completely reversible. We have been able to observe these same reversible red shifts in emission spectra by increasing the temperature of the reaction, by carrying out the reaction in 0.2 M urea and at normal pH values (7.6) in glycyl glycine buffer, by adding small concentrations of Zn++, Cd++, and Hg++ cations, as chlorides. The normalized emission spectra of the in vitro bioluminescence of purified Photinus pyralis luciferase for various Zn++ concentrations are shown in Figure 1. -

Bioluminescence in Insect
Int.J.Curr.Microbiol.App.Sci (2018) 7(3): 187-193 International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 7 Number 03 (2018) Journal homepage: http://www.ijcmas.com Review Article https://doi.org/10.20546/ijcmas.2018.703.022 Bioluminescence in Insect I. Yimjenjang Longkumer and Ram Kumar* Department of Entomology, Dr. Rajendra Prasad Central Agricultural University, Pusa, Bihar-848125, India *Corresponding author ABSTRACT Bioluminescence is defined as the emission of light from a living organism K e yw or ds that performs some biological function. Bioluminescence is one of the Fireflies, oldest fields of scientific study almost dating from the first written records Bioluminescence , of the ancient Greeks. This article describes the investigations of insect Luciferin luminescence and the crucial role imparted in the activities of insect. Many Article Info facets of this field are easily accessible for investigation without need for Accepted: advanced technology and so, within the History of Science, investigations 04 February 2018 of bioluminescence played a significant role in the establishment of the Available Online: scientific method, and also were among the many visual phenomena to be 10 March 2018 accounted for in developing a theory of light. Introduction Bioluminescence (BL) serves various purposes, including sexual attraction and When a living organism produces and emits courtship, predation and defense (Hastings and light as a result of a chemical reaction, the Wilson, 1976). This process is suggested to process is known as Bioluminescence - bio have arisen after O2 appearance on Earth at means 'living' in Greek while `lumen means least 30 different times during evolution, as 'light' in Latin. -

Brazilian Bioluminescent Beetles: Reflections on Catching Glimpses of Light in the Atlantic Forest and Cerrado
Anais da Academia Brasileira de Ciências (2018) 90(1 Suppl. 1): 663-679 (Annals of the Brazilian Academy of Sciences) Printed version ISSN 0001-3765 / Online version ISSN 1678-2690 http://dx.doi.org/10.1590/0001-3765201820170504 www.scielo.br/aabc | www.fb.com/aabcjournal Brazilian Bioluminescent Beetles: Reflections on Catching Glimpses of Light in the Atlantic Forest and Cerrado ETELVINO J.H. BECHARA and CASSIUS V. STEVANI Departamento de Química Fundamental, Instituto de Química, Universidade de São Paulo, Av. Prof. Lineu Prestes, 748, 05508-000 São Paulo, SP, Brazil Manuscript received on July 4, 2017; accepted for publication on August 11, 2017 ABSTRACT Bioluminescence - visible and cold light emission by living organisms - is a worldwide phenomenon, reported in terrestrial and marine environments since ancient times. Light emission from microorganisms, fungi, plants and animals may have arisen as an evolutionary response against oxygen toxicity and was appropriated for sexual attraction, predation, aposematism, and camouflage. Light emission results from the oxidation of a substrate, luciferin, by molecular oxygen, catalyzed by a luciferase, producing oxyluciferin in the excited singlet state, which decays to the ground state by fluorescence emission. Brazilian Atlantic forests and Cerrados are rich in luminescent beetles, which produce the same luciferin but slightly mutated luciferases, which result in distinct color emissions from green to red depending on the species. This review focuses on chemical and biological aspects of Brazilian luminescent beetles (Coleoptera) belonging to the Lampyridae (fireflies), Elateridae (click-beetles), and Phengodidae (railroad-worms) families. The ATP- dependent mechanism of bioluminescence, the role of luciferase tuning the color of light emission, the “luminous termite mounds” in Central Brazil, the cooperative roles of luciferase and superoxide dismutase against oxygen toxicity, and the hypothesis on the evolutionary origin of luciferases are highlighted. -

(12) United States Patent (10) Patent No.: US 9,574.223 B2 Cali Et Al
USO095.74223B2 (12) United States Patent (10) Patent No.: US 9,574.223 B2 Cali et al. (45) Date of Patent: *Feb. 21, 2017 (54) LUMINESCENCE-BASED METHODS AND 4,826,989 A 5/1989 Batz et al. PROBES FOR MEASURING CYTOCHROME 4,853,371 A 8/1989 Coy et al. 4,992,531 A 2f1991 Patroni et al. P450 ACTIVITY 5,035,999 A 7/1991 Geiger et al. 5,098,828 A 3/1992 Geiger et al. (71) Applicant: PROMEGA CORPORATION, 5,114,704 A 5/1992 Spanier et al. Madison, WI (US) 5,283,179 A 2, 1994 Wood 5,283,180 A 2f1994 Zomer et al. 5,290,684 A 3/1994 Kelly (72) Inventors: James J. Cali, Verona, WI (US); Dieter 5,374,534 A 12/1994 Zomer et al. Klaubert, Arroyo Grande, CA (US); 5,498.523 A 3, 1996 Tabor et al. William Daily, Santa Maria, CA (US); 5,641,641 A 6, 1997 Wood Samuel Kin Sang Ho, New Bedford, 5,650,135 A 7/1997 Contag et al. MA (US); Susan Frackman, Madison, 5,650,289 A T/1997 Wood 5,726,041 A 3/1998 Chrespi et al. WI (US); Erika Hawkins, Madison, WI 5,744,320 A 4/1998 Sherf et al. (US); Keith V. Wood, Mt. Horeb, WI 5,756.303 A 5/1998 Sato et al. (US) 5,780.287 A 7/1998 Kraus et al. 5,814,471 A 9, 1998 Wood (73) Assignee: PROMEGA CORPORATION, 5,876,946 A 3, 1999 Burbaum et al. 5,976,825 A 11/1999 Hochman Madison, WI (US) 6,143,492 A 11/2000 Makings et al. -

Study of the Photophysical Properties of Oxyluciferin Derivatives and Their Applications to the Characterization of Interactions Between Biomolecules Avisek Ghose
Study of the photophysical properties of oxyluciferin derivatives and their applications to the characterization of interactions between biomolecules Avisek Ghose To cite this version: Avisek Ghose. Study of the photophysical properties of oxyluciferin derivatives and their applica- tions to the characterization of interactions between biomolecules. Biological Physics [physics.bio-ph]. Université de Strasbourg, 2015. English. NNT : 2015STRAJ005. tel-01357655 HAL Id: tel-01357655 https://tel.archives-ouvertes.fr/tel-01357655 Submitted on 30 Aug 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITÉ DE STRASBOURG ÉCOLE DOCTORALE DES SCIENCES DE LA VIE ET DE LA SANTE (ED-414) UMR CNRS 7213, Faculté de Pharmacie, Illkirch THÈSE présentée par : Avisek GHOSE Soutenue le : 17 Février 2015 pour obtenir le grade de : !"#$%&'($')R%+,-$&.,# ' de Strasbourg Discipline/ Spécialité : Biophysique TITRE de la thèse Etude des propriétés photophysiques de dérivés ($')R!01)%",2 rine $#')$%&.'344),"3#,!+.'')R #%($' (R,+#$&3"#,!+.'$+#&$'6,!7!) "%)$. THÈSE dirigée par : M. ARNTZ Youri Maître de Conférences, Université de Strasbourg RAPPORTEURS : Mme. NAVIZET Isabelle Professeur des Universités, Université Paris-Est M. GUSTAVSSON Thomas Directeur de Recherche, CNRS AUTRES MEMBRES DU JURY : M. LEONARD Jérémie Chargé de Recherche, Université de Strasbourg M. -

Presentation1
Abstract In our search for multiplexed assays that increase both assay speed and sensitivity we have developed LiveResponseTM a panel of 5 secreted ultrasensitive luciferase reporters Gaussia luciferase (emission max 482 TM nm), Cypridina luciferase (emission max 463 nm), Blueshifted Renilla luciferase emission max 467 nm, Green LiveResponse A Panel of 5 Novel Ultra Renilla luciferase (emission max 527 nm) and a Red Italica firefly luciferase (emission max 613 nm). These significantly brighter luciferases (Gaussia luciferase is over a 1000fold brighter than the Photinus firefly luciferase and the redemitting Luciola luciferase is a 1000 times brighter than it's native counterpart) offer sensitive Secreted Luciferase Reporters for Cell increased sensitivity in screening applications involving analysis of weak promoters or hardtotransfect cells. The green variant of Renilla luciferase that is a component of the LiveResponseTM system has 100fold greater signal intensity and offers improved stability of the luminescent signal both in vitro and in vivo. This study demonstrates based HTS Applications the usefulness of these reporters to study multiple pathways within the same cell and profile responsiveness at Targeting different times (without cell lysis) using NFkB and CREB response elements to follow apoptosis and GPCR profiling in the same group of transfected cells. The LiveResponseTM system (a collection of 5 luciferase Systems reporters) and assay reagents for the same) offers flexibility in assay format i.e.. different reporters can be assayed individually using separate aliquots of the supernatant or up to three luciferases can be assayed in the Rampyari Raja Walia same sample of supernatant and the relative activities of the different luciferases can be spectrally resolved using appropriate filters. -

Firefly Luciferin
Firefly Luciferin Down the Rabbit Hole of Quirality (no Quantum Madness today…) Pathway inside Photinus pyralis Notice that while luciferin is a quiral protein, oxyluciferin is not. How to Recycle oxyluciferin: pathway Luc LRE No one How to Recycle oxyluciferin: enzyme Meet: Luciferine Regenerating Enzyme in Photinus pyralis ergo: • Sustainable reconversion reaction of oxyluciferin into D-luciferin. • Quirality of D-luciferin donated by D-cysteine. • With L-cysteine, oxyluciferin becomes L- luciferine. • However, only D-luciferin has luminescent properties! • Ergo: we need either D-cysteine or D-luciferin Enter Magic Enzyme • Magic Enzyme will stereoisomericaly bio- inverse L-luciferin into D-luciferin • Magic Enzyme will be simple and cheap • Magic Enzyme will be coded by a few genes • Magic Enzyme will be expressable in E. coli • Magic Enzyme won’t have complexities… • Does Magic Enzyme exist? A Voice from Beyond • “We previously reported that in the presence of ATP, Mg2+ and CoA, firefly luciferase exhibits coenzyme A ligase (CoA-ligase) activity on L-luciferin in vitro, but not on D- luciferin, to give luciferyl-CoA” Magic Quirality Conversion The Prestige • Magic enzyme is Luciferase! • “Lembert first reported the light production from L-luciferin and proposed that L-luciferin was racemized to give D-luciferin and it was effectively stimulated by the addition of pyrophosphate. This racemization is an in vitro reaction of luciferase in the absence of CoA. In the presence of CoA, L-luciferin is readily converted into luciferyl-CoA.” Huh?! • Initial medium inoculation with D-luciferin • Luciferase takes D-luciferin, produces oxyluciferin • Oxyluciferin is tranformed into L-luciferin via action of LRE with L-cysteine • L-luciferin is transformed into D-luciferin through Luciferase action So: • We CAN use Photinus pyralis luciferase • Suggested red-shifted mutant: Ser284Thr shines at 615nm Sources. -

Oxidation of Firefly Luciferin (Luminescence/Luciferase/Oxygen Exchange/Reaction Mechanism) OSAMU SHIMOMURA*, TOSHIO Gotot, and FRANK H
Proc. Nati. Acad. Scf. USA Vol. 74, No. 7, pp. 2799-2802, July 1977 Biochemistry Source of oxygen in the CO2 produced in the bioluminescent oxidation of firefly luciferin (luminescence/luciferase/oxygen exchange/reaction mechanism) OSAMU SHIMOMURA*, TOSHIO GOTOt, AND FRANK H. JOHNSON* * Department of Biology, Princeton University, Princeton, New Jersey 08540; and t Department of Agricultural Chemistry, Nagoya University, Chikusa, Nagoya 464, Japan Communicated by Henry Eyring, May 2, 1977 ABSTRACT Incorporation of 180 into the CO2 produced in luciferin by labeling product CO2 with 180 fully supported the bioluminescent oxidation of firefly luciferin was studied. scheme 1 (13-15). In regard to Renilla luciferin, however, one In H2160 medium with 1802 gas, the product CO2 contained up 0 of the was to arise from solvent to 75% C'60180, showing that one 0 of the product CO2 arose product CO2 reported H120 from the 02 that oxidized luciferin. This result is consistent with in both bioluminescence (3) and chemiluminescence (12), a dioxetane mechanism. Analysis of the mass spectral data of which would seem odd in view of the structural similarity be- the CO2 obtained in high-enrichment H2180 medium with 1602 tween Renilla luciferin and Cypridina luciferin. gas indicated the presence of about 20% contaminating CO2, The present study unambiguously supports scheme 1, but not which contributes approximately 70% of the total incorporated scheme 2, for the bioluminescence of firefly luciferin. More- 180. Thus the values of incorporated 180 in 112180 medium with over, present data cast a serious doubt on the validity of pre- 1802 gas have no significance in the present context. -

Illuminating Insights Into Firefly Luciferase and Other
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Chemistry & Biology Review Illuminating Insights into Firefly Luciferase and Other Bioluminescent Reporters Used in Chemical Biology Natasha Thorne,1 James Inglese,1 and Douglas S. Auld1,* 1NIH Chemical Genomics Center, National Institutes of Health, Bethesda, MD 20892-3370, USA *Correspondence: [email protected] DOI 10.1016/j.chembiol.2010.05.012 Understanding luciferase enzymology and the structure of compounds that modulate luciferase activity can be used to improve the design of luminescence-based assays. This review provides an overview of these popular reporters with an emphasis on the commonly used firefly luciferase from Photinus pyralis (FLuc). Large-scale chemical profile studies have identified a variety of scaffolds that inhibit FLuc. In some cell- based assays, these inhibitors can act in a counterintuitive way, leading to a gain in luminescent signal. Although formerly attributed to transcriptional activation, intracellular stabilization of FLuc is the primary mechanism underlying this observation. FLuc inhibition and stabilization can be complex, as illustrated by the compound PTC124, which is converted by FLuc in the presence of ATP to a high affinity multisubstrate adduct inhibitor, PTC124-AMP. The potential influence these findings can have on drug discovery efforts is provided here. Introduction of certain beetles known as glow worms (Phengodidae), and Technical advances in molecular biology, microtiter plate–based various marine organisms such as the sea pansy (Renillidae). spectrophotometry, and automation have enabled the develop- This review provides a summary of commonly used luciferases ment of biologically relevant assays with robust performance and suggests recommendations for their appropriate use in that can be utilized in high-throughput screening (HTS) for chem- common HTS assays.