Frawley Poster (NHRE 2016)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Japanese Maples – Acer Spp
Japanese Maples – Acer spp. Known for their astounding variety in color, texture and habit, Japanese Maples are easygoing and adaptable and belong in every garden. If you already have one, think about adding another! for every site, a japanese maple! • Botanists count twenty-three species to include under the common heading of Japanese Maple. The largest group is Acer palmatum and its cultivars, followed by Acer japonicum. The numerous cultivars of Acer palmatum are further divided into seven groups which are mostly defined by leaf-shape. • In addition to variation in leaf-shape, Japanese Maples come in an array of sizes, growth habits, color ranges, and full sun to part shade preference. They add structure, contrast, texture, and beauty to any garden. • Carefully evaluate your chosen planting spot: Japanese maples require a slightly acid, sandy loam, with medium moisture and good drainage. Regular leafed varieties take full sun better than laceleaf types, but all Japanese Maples will be happy with at least some afternoon shade, since St. Louis summer sun is hot, strong and humid. • Remember that plants grow outwards as well as upwards and site appropriately. Take into account proximity to any buildings and any overhead limits. Some pruning is possible but should not be a necessary task. • The main requirement of a Japanese Maple is a uniform supply of water. They are not happy with very wet periods followed by long, dry periods or vice versa. Most will need supplemental water in the St. Louis summers. Japanese Maples in planters have this same need for consistency – be careful that they do not become waterlogged! prepare your site, plant your tree • Japanese Maples are easy to plant. -

Junker's Nursery
1 JUNKER’S NURSERY LTD 2011-2012 Higher Cobhay (01823) 400075 Milverton Somerset E-mail: [email protected] TA4 1NJ Website: www.junker.co.uk See website for more detail: www.junker.co.uk elcome to a new look catalogue. Mind you, the catalogue is season, but if that means planting at a time of year when the plants have2 the least of the changes this year! We have finally completed best chance of success, then that can only be a good thing. I would en- W our relocation to our new site. Although we have owned it courage you therefore to make your plans and reserve your plants for for nearly 4 years now, personal circumstances intervened and it has when we can lift them. Typically we start this in mid-October. The taken longer to complete the move than we anticipated. It was definitely plants don’t need to have completely lost their leaves, but they do need worth the wait though (even if the rain is streaming down the windows to have finished active growth for the season. We will then continue lift- yet again, even as I write this in late July!) So much is different that it’s ing right through until March or so, but late planting can be risky in the difficult to know where to start...what remains unchanged however is our event of a dry spring such as we’ve had the last couple of years. Lifting commitment to growing the most exciting plants to the best of our abil- is weather dependant though, as we can’t continue if everything is water- ity, and to giving you, the customer, the kind of personal service that logged or frozen solid. -

Bigleaf Maple Decline in Western Washington
Bigleaf Maple Decline in Western Washington Jacob J. Betzen A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science University of Washington 2018 Committee: Patrick Tobin Gregory Ettl Brian Harvey Robert Harrison Program Authorized to Offer Degree: Environmental and Forest Sciences © Copyright 2018 Jacob Betzen University of Washington Abstract Bigleaf Maple Decline in Western Washington Jacob J. Betzen Chair of the Supervisory Committee: Professor Patrick Tobin School of Environmental and Forest Sciences Bigleaf maple (Acer macrophyllum Pursh) is a prominent component of the urban and suburban landscape in Western Washington, which lies at the heart of the native range of A. macrophyllum. Acer macrophyllum performs many important ecological, economic, and cultural functions, and its decline in the region could have cascading impacts. In 2011, increases in A. macrophyllum mortality were documented throughout the distributional range of the species. Symptoms of this decline included a systemic loss of vigor, loss of transpiration, and a reduction in photosynthetic potential, but did not display any signs or symptoms indicative of a specific causative agent. No pathogenic microbes, insects, or other biotic agents were initially implicated in causing or predisposing A. macrophyllum to decline. In my thesis research, I quantified the spatial extent and severity of A. macrophyllum decline in the urban, suburban, and wildland forests of western Washington, identified potential abiotic and biotic disturbance agents that are contributing to the decline, and conducted a dendrochronological analysis to ascertain the timing of the decline. I surveyed 22 sites that were previously reported as containing declining A. macrophyllum, and sampled 156 individual A. -

Non-Native Trees and Large Shrubs for the Washington, D.C. Area
Green Spring Gardens 4603 Green Spring Rd ● Alexandria ● VA 22312 Phone: 703-642-5173 ● TTY: 703-803-3354 www.fairfaxcounty.gov/parks/greenspring NON - NATIVE TREES AND LARGE SHRUBS FOR THE WASHINGTON, D.C. AREA Non-native trees are some of the most beloved plants in the landscape due to their beauty. In addition, these trees are grown for the shade, screening, structure, and landscape benefits they provide. Deciduous trees, whose leaves die and fall off in the autumn, are valuable additions to landscapes because of their changing interest throughout the year. Evergreen trees are valued for their year-round beauty and shelter for wildlife. Evergreens are often grouped into two categories, broadleaf evergreens and conifers. Broadleaf evergreens have broad, flat leaves. They also may have showy flowers, such as Camellia oleifera (a large shrub), or colorful fruits, such as Nellie R. Stevens holly. Coniferous evergreens either have needle-like foliage, such as the lacebark pine, or scale-like foliage, such as the green giant arborvitae. Conifers do not have true flowers or fruits but bear cones. Though most conifers are evergreen, exceptions exist. Dawn redwood, for example, loses its needles each fall. The following are useful definitions: Cultivar (cv.) - a cultivated variety designated by single quotes, such as ‘Autumn Gold’. A variety (var.) or subspecies (subsp.), in contrast, is found in nature and is a subdivision of a species (a variety of Cedar of Lebanon is listed). Full Shade - the amount of light under a dense deciduous tree canopy or beneath evergreens. Full Sun - at least 6 hours of sun daily. -

Acer Miyabei
Woody Plants Database [http://woodyplants.cals.cornell.edu] Species: Acer miyabei (ay'ser mi-YA-bee-eye) Miyabe Maple Cultivar Information * See specific cultivar notes on next page. Ornamental Characteristics Size: Tree < 30 feet Height: 35'-45', Width: 30' Leaves: Deciduous Shape: upright oval to rounded, can have open or dense branching, low branching Ornamental Other: prefers full sun, tolerates partial shade Environmental Characteristics Light: Full sun, Part shade Hardy To Zone: 5a Soil Ph: Can tolerate acid to alkaline soil (pH 5.0 to 8.0) CU Structural Soil™: Yes Insect Disease none of significance Bare Root Transplanting Easy Other easy to transplant B&B or < 2.5" caliper bare root. Native to Japan Moisture Tolerance 1 Woody Plants Database [http://woodyplants.cals.cornell.edu] Occasionally saturated Consistently moist, Occasional periods of Prolonged periods of or very wet soil well-drained soil dry soil dry soil 1 2 3 4 5 6 7 8 9 10 11 12 2 Woody Plants Database [http://woodyplants.cals.cornell.edu] Cultivars for Acer miyabei Showing 1-2 of 2 items. Cultivar Name Notes Rugged Ridge 'Rugged Ridge' - more deeply furrowed corky bark than species State Street 'State Street' (a.k.a. Morton) - hardy to zone 4; upright oval form; good uniform branching; dark green foliage; good golden yellow fall color; possibly fast growing 3 Woody Plants Database [http://woodyplants.cals.cornell.edu] Photos Acer miyabei trunk Acer miyabei foliage 4 Woody Plants Database [http://woodyplants.cals.cornell.edu] Acer miyabei habit Acer miyabei - Bark 5 Woody Plants Database [http://woodyplants.cals.cornell.edu] Acer miyabei - Leaf Acer miyabei - Habit 6. -

BIGLEAF MAPLE Evening Grosbeaks, Chipmunks, Mice, and a Variety of Birds
Plant Guide Wildlife: The seeds provide food for squirrels, BIGLEAF MAPLE evening grosbeaks, chipmunks, mice, and a variety of birds. Elk and deer browse the young twigs, leaves, Acer macrophyllum Pursh and saplings. Plant Symbol = ACMA3 Agroforestry: Bigleaf maple can be planted on sites Contributed By: USDA NRCS National Plant Data infected with laminated rot for site rehabilitation. It Center can also accelerate nutrient cycling, site productivity, revegetate disturbed riparian areas, and contribute to long-term sustainability. Status Please consult the PLANTS Web site and your State Department of Natural Resources for this plant’s current status, such as, state noxious status, and wetland indicator values. Description General: Maple Family (Aceraceae). Bigleaf maple is a native, long-lived medium to large sized deciduous tree that often grows to eighty feet tall. The leaves are simple, opposite, and very large between fifteen to thirty centimeters wide and almost as long (Farrar 1995). The flowers are yellow, Brother Alfred Brousseau © Saint Mary's College fragrant, and produced in noddling racemes @ CalPhotos appearing with the leaves in April or May. The fruit is paired, 2.5 - 4 centimeters long, and brown with Alternative Names stiff yellowish hair. The bark is smooth and gray- Oregon maple, broad leaf maple, big-leaf maple brown on young stems, becoming red-brown and deeply fissured, and broken into scales on the surface Uses (Preston 1989). Ethnobotanic: The inner bark was often dried and ground into a powder and then used as a thickener in Distribution: Acer macrophyllum is distributed soups or mixed with cereals when mixing bread. -

2. ACER Linnaeus, Sp. Pl. 2: 1054. 1753. 枫属 Feng Shu Trees Or Shrubs
Fl. China 11: 516–553. 2008. 2. ACER Linnaeus, Sp. Pl. 2: 1054. 1753. 枫属 feng shu Trees or shrubs. Leaves mostly simple and palmately lobed or at least palmately veined, in a few species pinnately veined and entire or toothed, or pinnately or palmately 3–5-foliolate. Inflorescence corymbiform or umbelliform, sometimes racemose or large paniculate. Sepals (4 or)5, rarely 6. Petals (4 or)5, rarely 6, seldom absent. Stamens (4 or 5 or)8(or 10 or 12); filaments distinct. Carpels 2; ovules (1 or)2 per locule. Fruit a winged schizocarp, commonly a double samara, usually 1-seeded; embryo oily or starchy, radicle elongate, cotyledons 2, green, flat or plicate; endosperm absent. 2n = 26. About 129 species: widespread in both temperate and tropical regions of N Africa, Asia, Europe, and Central and North America; 99 species (61 endemic, three introduced) in China. Acer lanceolatum Molliard (Bull. Soc. Bot. France 50: 134. 1903), described from Guangxi, is an uncertain species and is therefore not accepted here. The type specimen, in Berlin (B), has been destroyed. Up to now, no additional specimens have been found that could help clarify the application of this name. Worldwide, Japanese maples are famous for their autumn color, and there are over 400 cultivars. Also, many Chinese maple trees have beautiful autumn colors and have been cultivated widely in Chinese gardens, such as Acer buergerianum, A. davidii, A. duplicatoserratum, A. griseum, A. pictum, A. tataricum subsp. ginnala, A. triflorum, A. truncatum, and A. wilsonii. In winter, the snake-bark maples (A. davidii and its relatives) and paper-bark maple (A. -
![The Genus Acer (Maples) in Formosa and the Liukiu [Ryukyu] Islands](https://docslib.b-cdn.net/cover/3683/the-genus-acer-maples-in-formosa-and-the-liukiu-ryukyu-islands-593683.webp)
The Genus Acer (Maples) in Formosa and the Liukiu [Ryukyu] Islands
The Genus Acer (Maples) in Formosa and the Liukiu [Ryukyu] Islands H UI-LIN Ll1 THE SPECIES of the genus Aeer in Formosa C. Leaves glaucous beneath. and the Liukiu Islands are included in the D. Leaves obtuse or cuneate at base, revisional study of the family Aceraceae made not 3-nerved . 1. A . a/bopurpuraseens by Fang (1939). The Formosan species are DD. Leaves rounded to cordate and also treated by Kanehira in his work on the distinctly 3-nerved at base . Formosan trees (1936). The opinions ex . .. ... .. •2. A . itoanum pressed by these two authors are widely diver Cc. Leaves white-pubescent beneath . gent. Fang accepts practically all the species . .. .... ... .. 3. A. hypo/eueum originally described from Formosa, whereas BB. Leaves 3-lobed . Kanehira reduces a large number to synony .. .4. A. buergerianzon var. formosanum my . Neither of the two treatments is exhaus AA. Leaves serrate. tive, as a few names pertaining to Form osan B. Leaves undivided to shallowly 3 ~ .or plants are omitted from each . rarely 5-lobed. For purposes of the present study, the C. Leaves mostly undivided, sometimes works of these two authors, as well as other shallowly 3- or rarely 5-lobed; inflo pertinent literature, have been critically re rescence racemose. viewed . Specimens deposited in the U. S. D . Fruit 2-2.2 em. long . National Herbarium, Smithsonian Institution, . ... .. ...5. A. kawakamii and the herbarium of the National Taiwan DD. Fruit 2.5-3 em. long . University, Formosa, have been studied and . 5a. A . kawakamii vat. taiton arecited, with the abbreviations US and NTU, montanum respectively. -

Maples in the Landscape Sheriden Hansen, Jaydee Gunnell, and Andra Emmertson
EXTENSION.USU.EDU Maples in the Landscape Sheriden Hansen, JayDee Gunnell, and Andra Emmertson Introduction Maple trees (Acer sp.) are a common fixture and beautiful addition to Utah landscapes. There are over one hundred species, each with numerous cultivars (cultivated varieties) that are native to both North America and much of Northern Europe. Trees vary in size and shape, from small, almost prostrate forms like certain Japanese maples (Acer palmatum) and shrubby bigtooth maples (Acer grandidentatum) to large and stately shade trees like the Norway maple (Acer platanoides). Tree shape can vary greatly, ranging from upright, columnar, rounded, pyramidal to spreading. Because trees come in a Figure 1. Severe iron chlorosis on maple. Note the range of shapes and sizes, there is almost always a interveinal chlorosis characterized by the yellow leaves spot in a landscape that can be enhanced by the and green veins. Spotting on the leaves is indicative of the addition of a maple. Maples can create a focal point beginning of tissue necrosis from a chronic lack of iron. and ornamental interest in the landscape, providing interesting textures and colors, and of course, shade. some micronutrients, particularly iron, to be less Fall colors typically range from yellow to bright red, available, making it difficult for certain trees to take adding a burst of color to the landscape late in the up needed nutrients. A common problem associated season. with maples in the Intermountain West is iron chlorosis (Figure 1). This nutrient deficiency causes Recommended Cultivars yellowing leaves (chlorosis) with green veins, and in extreme conditions, can cause death of leaf edges. -
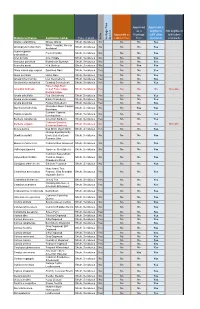
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Latitudinal Gradient in Leaf Defense Traits of Woody Plants Along Japanese Archipelago
Latitudinal gradient in leaf defense traits of woody plants along Japanese archipelago 日本産樹木種における、葉防御形質の緯度傾度 Saihanna 1 General Introduction It is estimated that over the twenty million species of organisms are living on our planet, and all of these organisms adapted to their own living environment, namely niche (Hatchinson 1957). Not only the abiotic factors but biotic interaction plays a key role in the maintenance of biodiversity. Animal-plant interactions are one of the most important topic in community ecology (e.g. Morin 1999). Plants and herbivore insects have accounted for about half of the entire diversity on the earth (Strong et al., 1984). Plant-herbivore interactions are extremely complex, which should lead the tremendous diversity of both plants and herbivores (e.g. Gutierrez et al., 1984; Hay et al., 1989). Although the interaction between these two components, namely co-speciation, should account for this diversification, most of the studies so far, tend to explain this interaction only from one side of them. Plants have interacted with insect herbivores for several hundred million years, which should lead to complex defense systems against various herbivores (Fürstenberg-Hägg et al., 2013). This interaction between plants and herbivores has long proposed the opportunity for studying the mechanism of the creation and maintenance of biological diversity because of its universality and generality (Strong et al. 1984; Ali and Agrawal 2012). It is believed that the evolution of plant defense traits followed by counter-adaptations in herbivores could lead to bursts of adaptive radiation of both components (Ehrlich and Raven 1969). Understanding the coevolution of plant and insect species and macroevolution of adaptive traits has inspired biologists for some decades, yet has been challenging to study even present days (Schluter, 2000). -

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m.