Differential Effects of Post-Weaning Diet and Maternal Obesity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Measurement of Metabolite Variations and Analysis of Related Gene Expression in Chinese Liquorice (Glycyrrhiza Uralensis) Plants
www.nature.com/scientificreports OPEN Measurement of metabolite variations and analysis of related gene expression in Chinese liquorice Received: 15 March 2017 Accepted: 28 March 2018 (Glycyrrhiza uralensis) plants under Published: xx xx xxxx UV-B irradiation Xiao Zhang1,2, Xiaoli Ding3,4, Yaxi Ji1,2, Shouchuang Wang5, Yingying Chen1,2, Jie Luo5, Yingbai Shen1,2 & Li Peng3,4 Plants respond to UV-B irradiation (280–315 nm wavelength) via elaborate metabolic regulatory mechanisms that help them adapt to this stress. To investigate the metabolic response of the medicinal herb Chinese liquorice (Glycyrrhiza uralensis) to UV-B irradiation, we performed liquid chromatography tandem mass spectrometry (LC-MS/MS)-based metabolomic analysis, combined with analysis of diferentially expressed genes in the leaves of plants exposed to UV-B irradiation at various time points. Fifty-four metabolites, primarily amino acids and favonoids, exhibited changes in levels after the UV-B treatment. The amino acid metabolism was altered by UV-B irradiation: the Asp family pathway was activated and closely correlated to Glu. Some amino acids appeared to be converted into antioxidants such as γ-aminobutyric acid and glutathione. Hierarchical clustering analysis revealed that various favonoids with characteristic groups were induced by UV-B. In particular, the levels of some ortho- dihydroxylated B-ring favonoids, which might function as scavengers of reactive oxygen species, increased in response to UV-B treatment. In general, unigenes encoding key enzymes involved in amino acid metabolism and favonoid biosynthesis were upregulated by UV-B irradiation. These fndings lay the foundation for further analysis of the mechanism underlying the response of G. -

Part One Amino Acids As Building Blocks
Part One Amino Acids as Building Blocks Amino Acids, Peptides and Proteins in Organic Chemistry. Vol.3 – Building Blocks, Catalysis and Coupling Chemistry. Edited by Andrew B. Hughes Copyright Ó 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim ISBN: 978-3-527-32102-5 j3 1 Amino Acid Biosynthesis Emily J. Parker and Andrew J. Pratt 1.1 Introduction The ribosomal synthesis of proteins utilizes a family of 20 a-amino acids that are universally coded by the translation machinery; in addition, two further a-amino acids, selenocysteine and pyrrolysine, are now believed to be incorporated into proteins via ribosomal synthesis in some organisms. More than 300 other amino acid residues have been identified in proteins, but most are of restricted distribution and produced via post-translational modification of the ubiquitous protein amino acids [1]. The ribosomally encoded a-amino acids described here ultimately derive from a-keto acids by a process corresponding to reductive amination. The most important biosynthetic distinction relates to whether appropriate carbon skeletons are pre-existing in basic metabolism or whether they have to be synthesized de novo and this division underpins the structure of this chapter. There are a small number of a-keto acids ubiquitously found in core metabolism, notably pyruvate (and a related 3-phosphoglycerate derivative from glycolysis), together with two components of the tricarboxylic acid cycle (TCA), oxaloacetate and a-ketoglutarate (a-KG). These building blocks ultimately provide the carbon skeletons for unbranched a-amino acids of three, four, and five carbons, respectively. a-Amino acids with shorter (glycine) or longer (lysine and pyrrolysine) straight chains are made by alternative pathways depending on the available raw materials. -

35 Disorders of Purine and Pyrimidine Metabolism
35 Disorders of Purine and Pyrimidine Metabolism Georges van den Berghe, M.- Françoise Vincent, Sandrine Marie 35.1 Inborn Errors of Purine Metabolism – 435 35.1.1 Phosphoribosyl Pyrophosphate Synthetase Superactivity – 435 35.1.2 Adenylosuccinase Deficiency – 436 35.1.3 AICA-Ribosiduria – 437 35.1.4 Muscle AMP Deaminase Deficiency – 437 35.1.5 Adenosine Deaminase Deficiency – 438 35.1.6 Adenosine Deaminase Superactivity – 439 35.1.7 Purine Nucleoside Phosphorylase Deficiency – 440 35.1.8 Xanthine Oxidase Deficiency – 440 35.1.9 Hypoxanthine-Guanine Phosphoribosyltransferase Deficiency – 441 35.1.10 Adenine Phosphoribosyltransferase Deficiency – 442 35.1.11 Deoxyguanosine Kinase Deficiency – 442 35.2 Inborn Errors of Pyrimidine Metabolism – 445 35.2.1 UMP Synthase Deficiency (Hereditary Orotic Aciduria) – 445 35.2.2 Dihydropyrimidine Dehydrogenase Deficiency – 445 35.2.3 Dihydropyrimidinase Deficiency – 446 35.2.4 Ureidopropionase Deficiency – 446 35.2.5 Pyrimidine 5’-Nucleotidase Deficiency – 446 35.2.6 Cytosolic 5’-Nucleotidase Superactivity – 447 35.2.7 Thymidine Phosphorylase Deficiency – 447 35.2.8 Thymidine Kinase Deficiency – 447 References – 447 434 Chapter 35 · Disorders of Purine and Pyrimidine Metabolism Purine Metabolism Purine nucleotides are essential cellular constituents 4 The catabolic pathway starts from GMP, IMP and which intervene in energy transfer, metabolic regula- AMP, and produces uric acid, a poorly soluble tion, and synthesis of DNA and RNA. Purine metabo- compound, which tends to crystallize once its lism can be divided into three pathways: plasma concentration surpasses 6.5–7 mg/dl (0.38– 4 The biosynthetic pathway, often termed de novo, 0.47 mmol/l). starts with the formation of phosphoribosyl pyro- 4 The salvage pathway utilizes the purine bases, gua- phosphate (PRPP) and leads to the synthesis of nine, hypoxanthine and adenine, which are pro- inosine monophosphate (IMP). -

Forms of Hypoxanthine Or, Through Interconversions, Guanine Or Adenine
826 GENETICS: GOTS AND GOLLUB PROC. N. A. S. Summary.-The ribonucleic acids of isolated thymus nuclei can be separated into two distinct fractions, one of which probably represents ribonucleic acid of the nu- cleolus. Studies of the incorporation of orotic acid-6-C"4 and adenosine-8-C'4 into these RNA fractions in vitro show great differences in their metabolic activity and different susceptibilities to an inhibitor of RNA synthesis, the "nucleolar" RNA being by far the more active. It is a pleasure to acknowledge our indebtedness to Mr. Rudolf Meudt for his careful and expert technical assistance. * This research was supported in part by a grant (RG-4919 M&G) from the United States Public Health Service. I V. G. Allfrey, A. E. Mirsky, and S. Osawa, Nature, 176, 1042, 1955. 2 V. G. Allfrey, A. E. Mirsky, and S. Osawa, J. Gen. Physiol., 40, 451, 1957. 3 R. Logan and J. N. Davidson, Biochim. et Biophys. Acta, 24, 196, 1957. 4 S. Osawa, K. Takata, and Y. Hotta (in press). 6 J. M. Webb, J. Biol. Chem., 221, 635, 1956. 6 M. Bessis, in Traite de cytologie sanguine (Paris: Masson & Cie, 1954), p. 83. J. N. Davidson and R. M. S. Smellie. Biochem. J.. 52, 594, 1952. SEQUENTIAL BLOCKADE IN ADENINE BIOSYNTHESIS BY GENETIC LOSS OF AN APPARENT BIFUNCTIONAL DEACYLASE* By JOSEPH S. GOTS AND EDITH G. GOLLUB DEPARTMENT OF MICROBIOLOGY, SCHOOL OF MEDICINE, UNIVERSITY OF PENNSYLVANIA, PHILADELPHIA, PENNSYLVANIA Communicated by D. Wright Wilson, July 3, 1957 Along the biosynthetic pathway to the purines of nucleic acids, inosinic acid occurs as a pivotal point in a bifurcation which leads to adenylic acid along one branch and to guanylic acid along the other: (B) r- Adenylic acid (AMP) (A) Inosinic acid (IMP) (C) L|_ * Guanylic acid (GMP) Bacterial mutations may result in genetic impairments at three locations with respect to the pivotal inosinic acid, thus yielding auxotrophs with three broad classes of nutritional response. -

Figure S1. Heat Map of R (Pearson's Correlation Coefficient)
Figure S1. Heat map of r (Pearson’s correlation coefficient) value among different samples including replicates. The color represented the r value. Figure S2. Distributions of accumulation profiles of lipids, nucleotides, and vitamins detected by widely-targeted UPLC-MC during four fruit developmental stages. The colors indicate the proportional content of each identified metabolites as determined by the average peak response area with R scale normalization. PS1, 2, 3, and 4 represents fruit samples collected at 27, 84, 125, 165 Days After Anthesis (DAA), respectively. Three independent replicates were performed for each stages. Figure S3. Differential metabolites of PS2 vs PS1 group in flavonoid biosynthesis pathway. Figure S4. Differential metabolites of PS2 vs PS1 group in phenylpropanoid biosynthesis pathway. Figure S5. Differential metabolites of PS3 vs PS2 group in flavonoid biosynthesis pathway. Figure S6. Differential metabolites of PS3 vs PS2 group in phenylpropanoid biosynthesis pathway. Figure S7. Differential metabolites of PS4 vs PS3 group in biosynthesis of phenylpropanoids pathway. Figure S8. Differential metabolites of PS2 vs PS1 group in flavonoid biosynthesis pathway and phenylpropanoid biosynthesis pathway combined with RNA-seq results. Table S1. A total of 462 detected metabolites in this study and their peak response areas along the developmental stages of apple fruit. mix0 mix0 mix0 Index Compounds Class PS1a PS1b PS1c PS2a PS2b PS2c PS3a PS3b PS3c PS4a PS4b PS4c ID 1 2 3 Alcohols and 5.25E 7.57E 5.27E 4.24E 5.20E -

COMPILATION of AMINO ACIDS, DRUGS, METABOLITES and OTHER COMPOUNDS in MASSTRAK AMINO ACID ANALYSIS SOLUTION Paula Hong, Kendon S
COMPILATION OF AMINO ACIDS, DRUGS, METABOLITES AND OTHER COMPOUNDS IN MASSTRAK AMINO ACID ANALYSIS SOLUTION Paula Hong, Kendon S. Graham, Alexandre Paccou, T homas E. Wheat and Diane M. Diehl INTRODUCTION LC conditions Physiological amino acid analysis is commonly performed to LC System: Waters ACQUITY UPLC® System with TUV monitor and study a wide variety of metabolic processes. A wide Column: MassTrak AAA Column 2.1 x 150 mm, 1.7 µm variety of drugs, foods, and metabolic intermediates that may Column Temp: 43 ˚C be present in biological fluids can appear as peaks in amino Flow Rate: 400 µL/min. acid analysis, therefore, it is important to be able to identify Mobile Phase A: MassTrak AAA Eluent A Concentrate, unknown compounds.1,2,3 The reproducibility and robustness of diluted 1:10 the MassTrak Amino Acid Analysis Solution make this method well Mobile Phase B: MassTrak AAA Eluent B suited to such a study as well.4 Weak Needle Wash: 5/95 Acetonitrile/Water Strong Needle Wash: 95/5 Acetonitrile/Water Gradient: MassTrak AAA Standard Gradient (as provided in kit) Detection: UV @ 260 nm Injection Volume: 1 µL EXPERIMENTAL Injection Mode: Partial Loop with Needle Overfill (PLNO) Compound sample preparation A library of compounds was assembled. Each compound was derivatized individually and spiked into the MassTrak™ AAA Solution Standard prior to chromatographic analysis. The elution RESULTS AND DISCUSSION position of each tested compound could be related to known amino acids. A wide variety of antibiotics, pharmaceutical compounds and metabolite by-products are found in biological fluids. The reten- 1. -

Impacts of Dietary Exposure to Pesticides on Faecal Microbiome
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448511; this version posted June 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Impacts of dietary exposure to pesticides on faecal microbiome 2 metabolism in adult twins 3 4 Robin Mesnage1, Ruth C E Bowyer2, Souleiman El Balkhi3, Franck Saint-Marcoux3, Arnaud 5 Gardere3, Quinten Raymond Ducarmon4, Anoecim Robecca Geelen4, Romy Daniëlle Zwittink4, 6 Dimitris Tsoukalas5, Evangelia Sarandi5, Efstathia I. Paramera6, Timothy Spector2, Claire J Steves2, 7 Michael N Antoniou1* 8 9 1 Gene Expression and Therapy Group, King's College London, Faculty of Life Sciences & Medicine, 10 Department of Medical and Molecular Genetics, Guy's Hospital, London, SE1 9RT, UK. 11 12 2 Department of Twin Research and Genetic Epidemiology, Kings College London, London, UK 13 14 3 Service de pharmacologie, toxicologie et pharmacovigilance, UF Toxicologie analytique environnementale 15 et santé au travail, CHU de Limoges, Limoges, France 16 17 4 Center for Microbiome Analyses and Therapeutics, Leiden University Medical Center, Leiden, The 18 Netherlands 19 20 5 Metabolomic Medicine Clinic, Health Clinics for Autoimmune and Chronic Diseases, 10674 Athens, 21 Greece 22 23 6 NEOLAB S.A., Medical laboratory, 125 Michalakopoulu Str., 11527 Athens, Greece 24 25 26 *Correspondence: [email protected] 27 bioRxiv preprint doi: https://doi.org/10.1101/2021.06.16.448511; this version posted June 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Intestinal Phosphate Transport in Familial ~~~O~Hos~Haternicrickets
FAMILIAL HYPOPHOSPHATEMIC RICKETS 69 1 pine oxidoreductase. The deficiencies were confirmed in skin Glutaric aciduria: A "new" disorder of amino acid metabolism. Biochem. fibroblasts from two siblings with the disease and a third patient Med., 12: 12 (1975). 15. Hutzler, J., and Dancis. J.: Saccharopine cleavage by a dehydrogenase of human from an unrelated family. liver. Biochim. Biophys. Acta. 206: 205 (1970). REFERENCES AND NOTES 16. Hutzler. J., and Dancis. J.: Preparative synthesis of saccharopine. Biochim. Biophys. Acta, 222: 225 (1970). I. Ampola. M. G., Ampola. M., Mian, M., Shaw, W., Levy, H., Letsou, A,, and 17. Hutzler, J., and Dancis, J.: Lysine-ketoglutarate reductase in human tissues. Doyle. M.: In preparation. Biochim. Biophys. Acta, 377: 42 (1975). 2. Bowden, J. A., and Connelly, J. L.: Branched-chain a-keto acid metabolism. I. 18. Krooth, R. S.: Constitutive mutations and hereditary enzyme deficiencies in Isolation, purification and partial characterization of bovine liver a- mammalian cells. In: M. Harris and B. Thompson: Regulation of Gene ketoisocaproic:a-keto-@-methylvaleric acid dehydrogenase. J. Biol. Chem., Expression in Eukaryotic Cells, p. 115 (Fogarty International Center, Be- 243: 1 198 (1968). thesda, Md., 1973). 3. Cox. R. P.. Krauss, M. R., Balis, M. E., and Dancis, J.: Communication between 19. Lormans, S., and Lowenthal. A,: Amino adipic aciduria in an oligophrenic child. normal and enzyme deficient cells in tissue culture. Exp. Cell Res.. 74: 251 Clin. Chim. Acta. 57: 97 (1974). (1972). 20. Lowry. 0. H., Rosehrough, N. J.. Farr. A. L., and Randall, R. J.: Protein 4. Cox. R. P., and MacLeod, C. -

Amino Acid Degradation
BI/CH 422/622 OUTLINE: OUTLINE: Protein Degradation (Catabolism) Digestion Amino-Acid Degradation Inside of cells Protein turnover Dealing with the carbon Ubiquitin Fates of the 29 Activation-E1 Seven Families Conjugation-E2 nitrogen atoms in 20 1. ADENQ Ligation-E3 AA: Proteosome 2. RPH 9 ammonia oxidase Amino-Acid Degradation 18 transamination Ammonia 2 urea one-carbon metabolism free transamination-mechanism to know THF Urea Cycle – dealing with the nitrogen SAM 5 Steps Carbamoyl-phosphate synthetase 3. GSC Ornithine transcarbamylase PLP uses Arginino-succinate synthetase Arginino-succinase 4. MT – one carbon metabolism Arginase 5. FY – oxidase vs oxygenase Energetics Urea Bi-cycle 6. KW – Urea Cycle – dealing with the nitrogen 7. BCAA – VIL Feeding the Urea Cycle Glucose-Alanine Cycle Convergence with Fatty acid-odd chain Free Ammonia Overview Glutamine Glutamate dehydrogenase Overall energetics Amino Acid A. Concepts 1. ConvergentDegradation 2. ketogenic/glucogenic 3. Reactions seen before The SEVEN (7) Families B. Transaminase (A,D,E) / Deaminase (Q,N) Family C. Related to biosynthesis (R,P,H; C,G,S; M,T) 1.Glu Family a. Introduce oxidases/oxygenases b. Introduce one-carbon metabolism (1C) 2.Pyruvate Family a. PLP reactions 3. a-Ketobutyric Family (M,T) a. 1-C metabolism D. Dedicated 1. Aromatic Family (F,Y) a. oxidases/oxygenases 2. a-Ketoadipic Family (K,W) 3. Branched-chain Family (V,I,L) E. Convergence with Fatty Acids: propionyl-CoA 29 N 1 Amino Acid Degradation • Intermediates of the central metabolic pathway • Some amino acids result in more than one intermediate. • Ketogenic amino acids can be converted to ketone bodies. -

Omega-3 Fatty Acids Correlate with Gut Microbiome Diversity And
Omega-3 fatty acids correlate with gut microbiome diversity and production of N- carbamylglutamate in middle aged and elderly women Cristina Mennia, Jonas Zierera, Tess Pallistera, Matthew A Jacksona, Tao Longb, Robert P Mohneyc, Claire J Stevesa, Tim D Spectora, Ana M Valdesa,d,e a Department of Twin Research and Genetic Epidemiology, Kings College London, London, UK. b Sanford Burnham Prebys, USA cMetabolon Inc., Raleigh-Durham, NC 27709, USA. dSchool of Medicine , Nottingham City Hospital, Hucknall Road, Nottingham, UK. e NIHR Nottingham Biomedical Research Centre, Nottingham, UK. Corresponding author: Dr Ana M Valdes School of Medicine Clinical Sciences Building, Nottingham City Hospital, Hucknall Road, Nottingham, NG5 1PB, UK Phone number: +44 (0)115 823 1954; Fax number:+44(0) 115 823 1757 email: [email protected] Supplementary Table 1. List of faecal metabolites significantly associated to DHA circulating levels (FDR<0.05)adjusting for age, BMI and family relatedness. Super Pathway Sub pathway Metabolite BETA SE P Q Lipid Polyunsaturated Fatty Acid (n3 and n6) eicosapentaenoate (EPA; 20:5n3) 0.15 0.04 4.35x10-5 0.01 Xenobiotics Carbamylated aminoacid N-carbamylglutamate 0.15 0.04 1.21x10-4 0.02 Peptide Dipeptide Derivative anserine 0.13 0.04 5.39x10-4 0.04 Supplementary Table 2. Associations between five measures of microbiome diversity and circulating levels of PUFA adjusting for age, BMI fibre intake and family relatedness SHANNON DIVERSITY CHAO1 OBSERVED Phylogenetic Diversity SIMPSON SPECIES Beta SE P Beta SE P Beta SE P Beta SE P Beta SE P DHA 0.13 0.04 0.002 0.12 0.04 0.003 0.14 0.04 0.001 0.12 0.04 0.01 0.09 0.04 0.02 FAW3 0.12 0.04 0.005 0.13 0.04 0.003 0.13 0.04 0.002 0.12 0.05 0.01 0.08 0.04 0.05 LA 0.10 0.05 0.02 0.10 0.05 0.03 0.10 0.05 0.03 0.10 0.05 0.03 0.09 0.04 0.04 (18:2) FAW6 0.09 0.04 0.02 0.10 0.04 0.01 0.10 0.04 0.01 0.10 0.04 0.02 0.08 0.04 0.05 DHA=22:6 docosahexaenoic acid;, FAW3= Omega-3 fatty acids; 18:2, LA= linoleic acid;; FAW6=omega-6 fatty acids Supplementary Table 3. -

X-Ray Fluorescence Analysis Method Röntgenfluoreszenz-Analyseverfahren Procédé D’Analyse Par Rayons X Fluorescents
(19) & (11) EP 2 084 519 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: G01N 23/223 (2006.01) G01T 1/36 (2006.01) 01.08.2012 Bulletin 2012/31 C12Q 1/00 (2006.01) (21) Application number: 07874491.9 (86) International application number: PCT/US2007/021888 (22) Date of filing: 10.10.2007 (87) International publication number: WO 2008/127291 (23.10.2008 Gazette 2008/43) (54) X-RAY FLUORESCENCE ANALYSIS METHOD RÖNTGENFLUORESZENZ-ANALYSEVERFAHREN PROCÉDÉ D’ANALYSE PAR RAYONS X FLUORESCENTS (84) Designated Contracting States: • BURRELL, Anthony, K. AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Los Alamos, NM 87544 (US) HU IE IS IT LI LT LU LV MC MT NL PL PT RO SE SI SK TR (74) Representative: Albrecht, Thomas Kraus & Weisert (30) Priority: 10.10.2006 US 850594 P Patent- und Rechtsanwälte Thomas-Wimmer-Ring 15 (43) Date of publication of application: 80539 München (DE) 05.08.2009 Bulletin 2009/32 (56) References cited: (60) Divisional application: JP-A- 2001 289 802 US-A1- 2003 027 129 12164870.3 US-A1- 2003 027 129 US-A1- 2004 004 183 US-A1- 2004 017 884 US-A1- 2004 017 884 (73) Proprietors: US-A1- 2004 093 526 US-A1- 2004 235 059 • Los Alamos National Security, LLC US-A1- 2004 235 059 US-A1- 2005 011 818 Los Alamos, NM 87545 (US) US-A1- 2005 011 818 US-B1- 6 329 209 • Caldera Pharmaceuticals, INC. US-B2- 6 719 147 Los Alamos, NM 87544 (US) • GOLDIN E M ET AL: "Quantitation of antibody (72) Inventors: binding to cell surface antigens by X-ray • BIRNBAUM, Eva, R. -
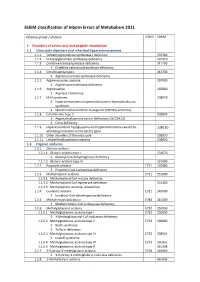
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2.